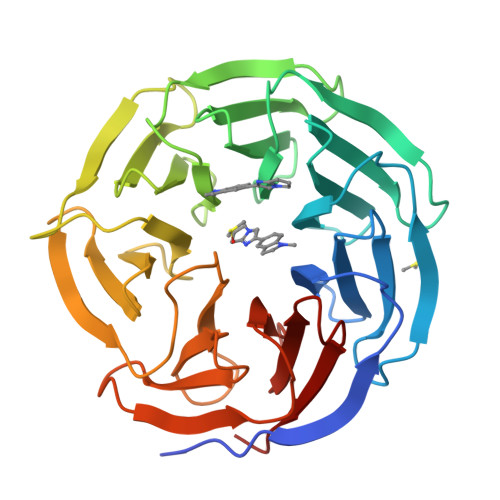
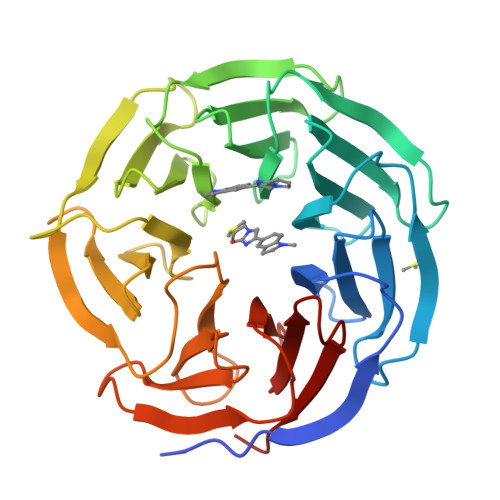
Discovery of Potent 2-Aryl-6,7-dihydro-5 H-pyrrolo[1,2- a]imidazoles as WDR5-WIN-Site Inhibitors Using Fragment-Based Methods and Structure-Based Design.
Wang, F., Jeon, K.O., Salovich, J.M., Macdonald, J.D., Alvarado, J., Gogliotti, R.D., Phan, J., Olejniczak, E.T., Sun, Q., Wang, S., Camper, D., Yuh, J.P., Shaw, J.G., Sai, J., Rossanese, O.W., Tansey, W.P., Stauffer, S.R., Fesik, S.W.(2018) J Med Chem 61: 5623-5642
- PubMed: 29889518
- DOI: https://doi.org/10.1021/acs.jmedchem.8b00375
- Primary Citation of Related Structures:
6D9X, 6DAI, 6DAK, 6DAR, 6DAS - PubMed Abstract:
WDR5 is a chromatin-regulatory scaffold protein overexpressed in various cancers and a potential epigenetic drug target for the treatment of mixed-lineage leukemia. Here, we describe the discovery of potent and selective WDR5-WIN-site inhibitors using fragment-based methods and structure-based design. NMR-based screening of a large fragment library identified several chemically distinct hit series that bind to the WIN site within WDR5. Members of a 6,7-dihydro-5 H-pyrrolo[1,2- a]imidazole fragment class were expanded using a structure-based design approach to arrive at lead compounds with dissociation constants <10 nM and micromolar cellular activity against an AML-leukemia cell line. These compounds represent starting points for the discovery of clinically useful WDR5 inhibitors for the treatment of cancer.