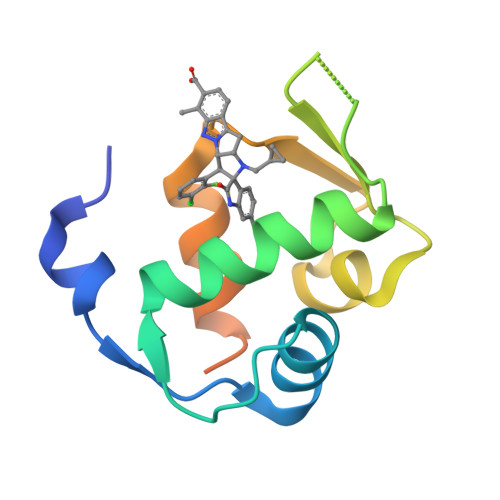
Discovery and Characterization of Brigimadlin, a Novel and Highly Potent MDM2-p53 Antagonist Suitable for Intermittent Dose Schedules.
Gollner, A., Rudolph, D., Weyer-Czernilofsky, U., Baumgartinger, R., Jung, P., Weinstabl, H., Ramharter, J., Grempler, R., Quant, J., Rinnenthal, J., Perez Pitarch, A., Golubovic, B., Gerlach, D., Bader, G., Wetzel, K., Otto, S., Mandl, C., Boehmelt, G., McConnell, D.B., Kraut, N., Sini, P.(2024) Mol Cancer Ther 23: 1689-1702
- PubMed: 39259562
- DOI: https://doi.org/10.1158/1535-7163.MCT-23-0783
- Primary Citation of Related Structures:
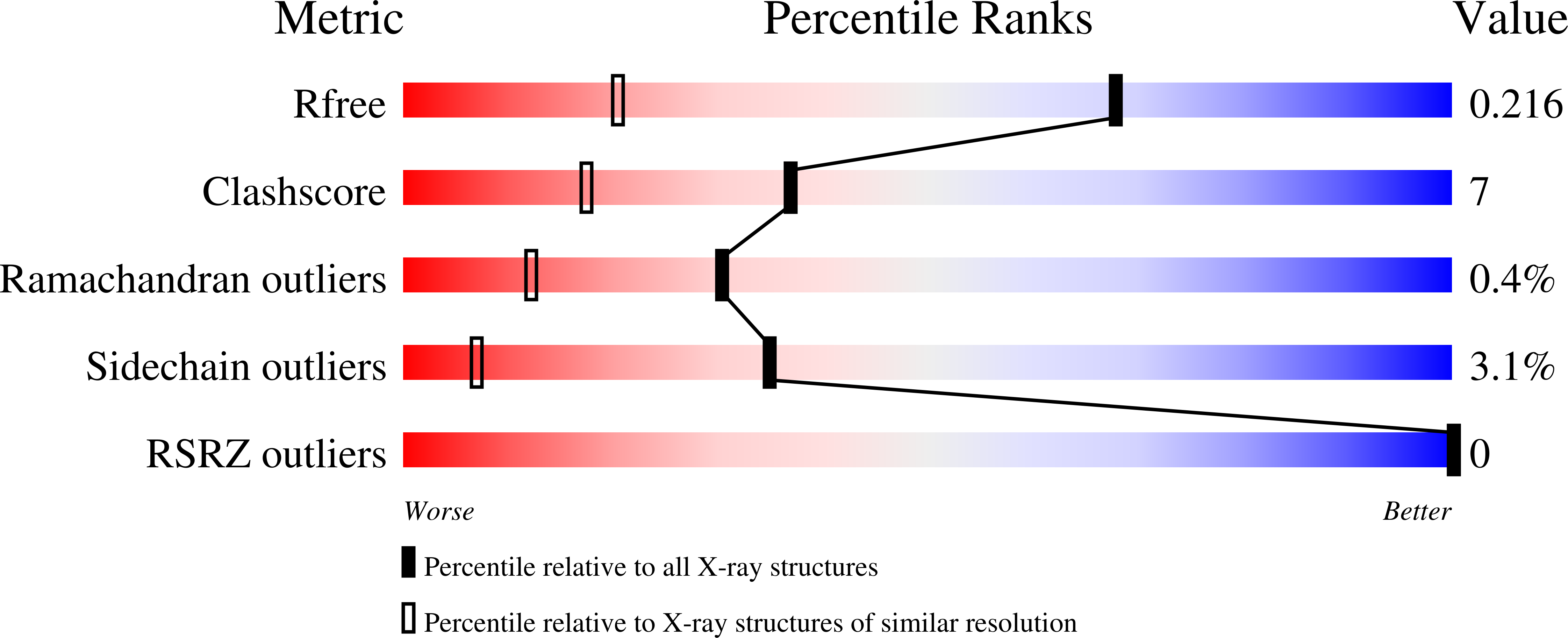
8PWC - PubMed Abstract:
p53 is known as the guardian of the genome and is one of the most important tumor-suppressors. It is inactivated in most tumors, either via tumor protein p53 (TP53) gene mutation or copy number amplification of key negative regulators, e.g., mouse double minute 2 (MDM2). Compounds that bind to the MDM2 protein and disrupt its interaction with p53 restore p53 tumor suppressor activity, thereby promoting cell cycle arrest and apoptosis. Previous clinical experience with MDM2-p53 protein-protein interaction antagonists (MDM2-p53 antagonists) have demonstrated that thrombocytopenia and neutropenia represent on-target dose-limiting toxicities that might restrict their therapeutic utility. Dosing less frequently, while maintaining efficacious exposure, represents an approach to mitigate toxicity and improve the therapeutic window of MDM2-p53 antagonists. However, to achieve this, a molecule possessing excellent potency and ideal pharmacokinetic properties is required. Here, we present the discovery and characterization of brigimadlin (BI 907828), a novel, investigational spiro-oxindole MDM2-p53 antagonist. Brigimadlin exhibited high bioavailability and exposure, as well as dose-linear pharmacokinetics in preclinical models. Brigimadlin treatment restored p53 activity and led to apoptosis induction in preclinical models of TP53 wild-type, MDM2-amplified cancer. Oral administration of brigimadlin in an intermittent dosing schedule induced potent tumor growth inhibition in several TP53 wild-type, MDM2-amplified xenograft models. Exploratory clinical pharmacokinetic studies (NCT03449381) showed high systemic exposure and a long plasma elimination half-life in cancer patients who received oral brigimadlin. These findings support the continued clinical evaluation of brigimadlin in patients with MDM2-amplified cancers, such as dedifferentiated liposarcoma.
Organizational Affiliation:
Boehringer Ingelheim RCV GmbH & Co KG, Vienna, Austria.