Structural Basis for the Essential Role of Ca 2+ in the Lytic Activity of Staphylococcus aureus PlyGRCS Endolysin Targeting Methicillin-Resistant Staphylococcus aureus.
Krishnappa, G., Nagaraj, H., SureshKumar, H.B., Mandal, M., Padavattan, S., Bahubali, V.H., Thiyagarajan, S., Padmanabhan, B.(2025) Proteins 93: 920-933
- PubMed: 39660753
- DOI: https://doi.org/10.1002/prot.26777
- Primary Citation of Related Structures:
8XYF - PubMed Abstract:
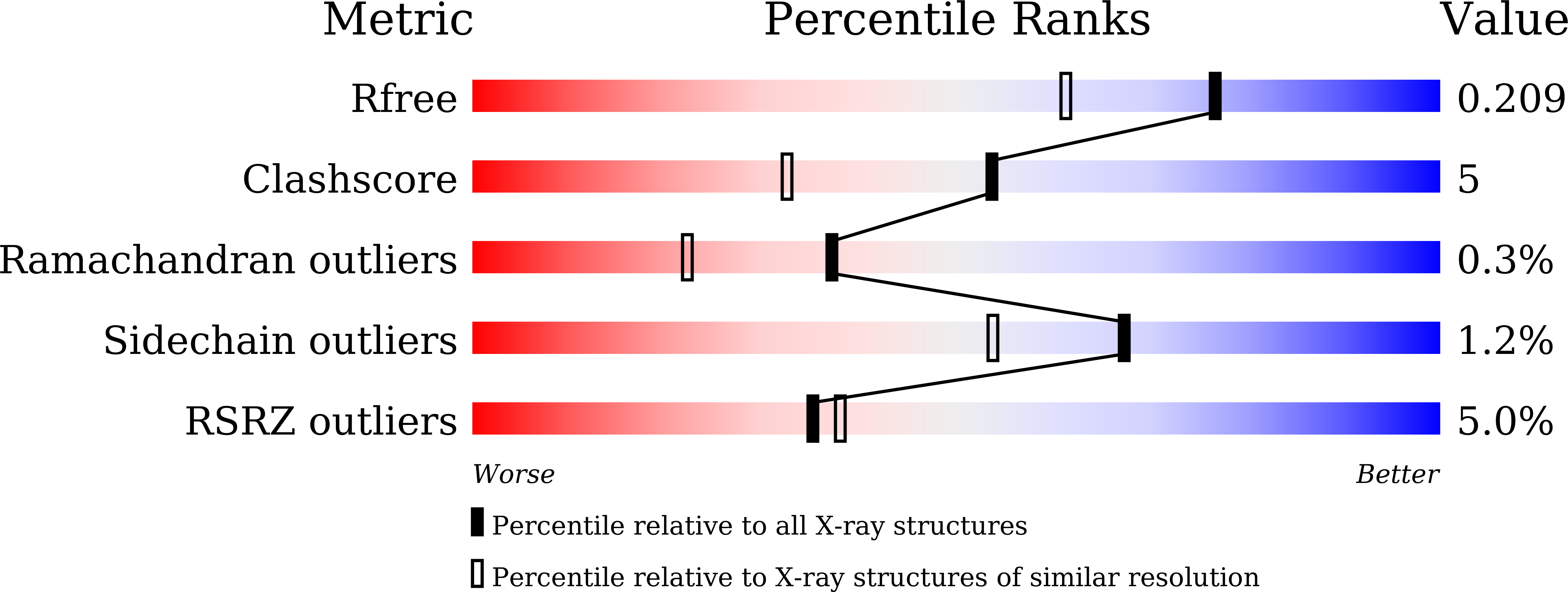
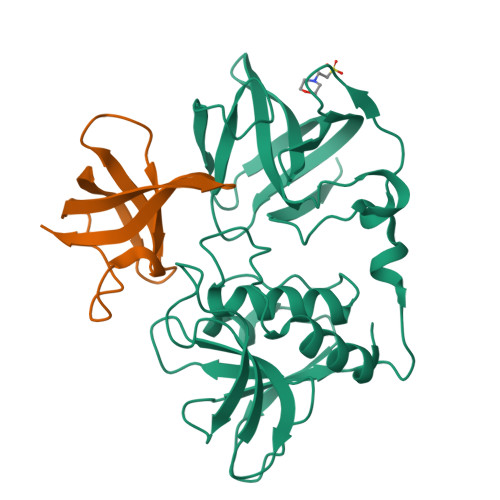
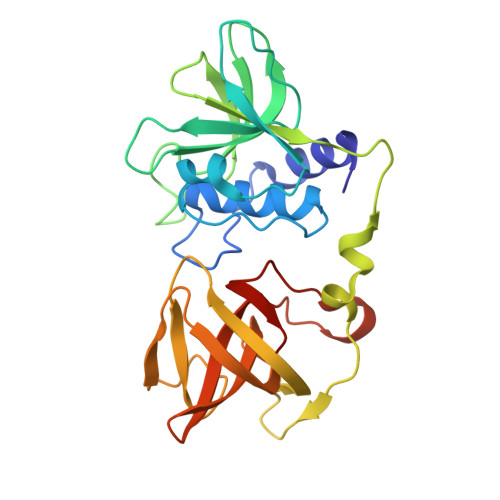
Staphylococcus aureus causes a wide range of infections, from mild skin conditions to severe, life-threatening diseases. Bacteriophage endolysins exhibit a selective capacity to degrade the peptidoglycan layer of Gram-positive bacteria, making promising biotherapeutic agents against antibiotic-resistant infections. PlyGRCS, a specific endolysin derived from S. aureus, comprises a catalytic CHAP domain and a cell-wall binding SH3_5 domain connected by a linker. Ca 2+ ions are essential for the CHAP domain's catalytic function. The crystal structure of PlyGRCS, determined in the absence of Ca 2+ and refined to a resolution of 1.67 Å, revealed significant conformational changes in the Ca 2+ binding site. Antimicrobial assays with Ca 2+ -deficient PlyGRCS and mutants targeting key residues in the catalytic and Ca 2+ binding regions highlighted the importance of specific functional residues for lytic activity against methicillin-resistant Staphylococcus aureus (MRSA). These structural and microbial studies provide valuable insights into the critical residues contributing to PlyGRCS's bacteriolytic efficacy against MRSA.
Organizational Affiliation:
Department of Biophysics, National Institute of Mental Health and Neurosciences (NIMHANS), Bengaluru, India.