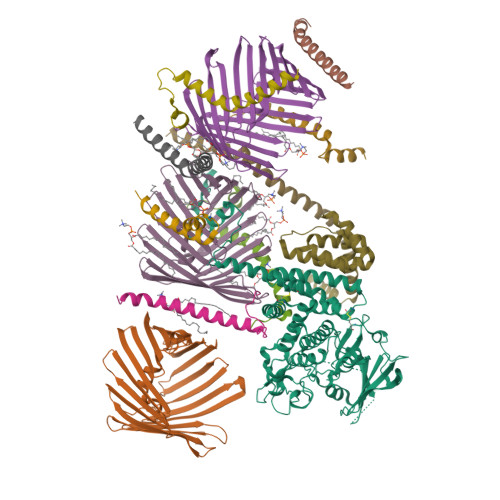
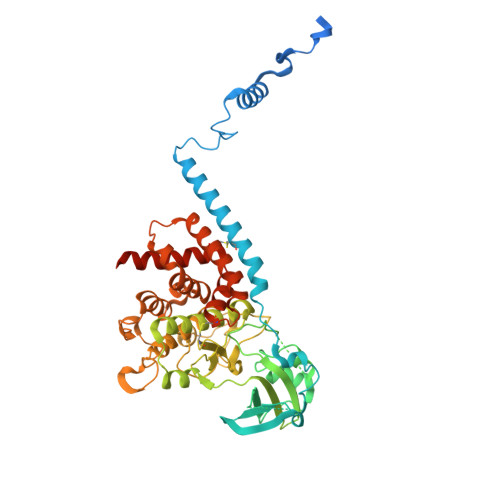
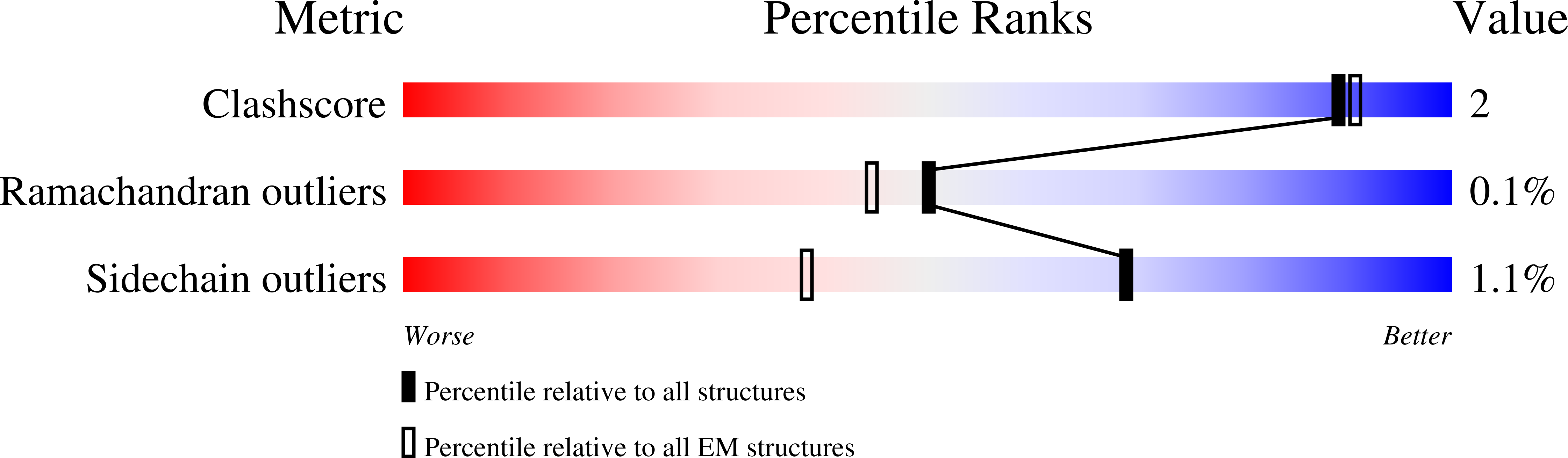Structure of human PINK1 at a mitochondrial TOM-VDAC array.
Callegari, S., Kirk, N.S., Gan, Z.Y., Dite, T., Cobbold, S.A., Leis, A., Dagley, L.F., Glukhova, A., Komander, D.(2025) Science : eadu6445-eadu6445
- PubMed: 40080546
- DOI: https://doi.org/10.1126/science.adu6445
- Primary Citation of Related Structures:
9EIH, 9EII, 9EIJ - PubMed Abstract:
Mutations in the ubiquitin kinase PINK1 cause early onset Parkinson's Disease, but how PINK1 is stabilized at depolarized mitochondrial translocase complexes has remained poorly understood. We determined a 3.1-Å resolution cryo-electron microscopy structure of dimeric human PINK1 stabilized at an endogenous array of mitochondrial TOM and VDAC complexes. Symmetric arrangement of two TOM core complexes around a central VDAC2 dimer is facilitated by TOM5 and TOM20, both of which also bind PINK1 kinase C-lobes. PINK1 enters mitochondria through the proximal TOM40 barrel of the TOM core complex, guided by TOM7 and TOM22. Our structure explains how human PINK1 is stabilized at the TOM complex and regulated by oxidation, uncovers a previously unknown TOM-VDAC assembly, and reveals how a physiological substrate traverses TOM40 during translocation.
Organizational Affiliation:
Walter and Eliza Hall Institute of Medical Research, Parkville, Victoria, Australia.