2009 PDB News
Contents:
Earlier news is available and is archived in the RCSB PDB newsletters.
15-December-2009
CCDC Archives 500,000th Crystal Structure
The Cambridge Crystallographic Data Centre (CCDC) has announced an important milestone in the history of crystallography--the archiving of the 500,000th small molecule crystal structure to the Cambridge Structural Database (CSD). For more information, visit the CCDC at www.ccdc.cam.ac.uk.
Use Widgets to Embed RCSB PDB Features on Your Website
Molecule of the Month images and text, static images and interactive views for any PDB structure, and RCSB PDB searches based on ID, keyword, or author name can be embedded into any website using the RCSB PDB's web widgets. These small bits of code can be customized to display and link to RCSB PDB features.
*
The RCSB PDB Molecule of the Month widget embeds an image from the feature and links to the entire article. The widget can be customized to specify width, colors, amount of text shown, and molecule.
The 'simple' widget version displaying the May 2003 Molecule of the Month
*
The Tag Library can be used to embed an image that links to the corresponding Structure Summary page; provide a menu of links to a particular entry's Jmol view, Structure Summary Page, and PDB file; and provide a link that performs a current keyword query.
Example text marked up using the Tag Library (from doi: 10.2210/rcsb_pdb/mom_2009_1)
* The RCSB PDB Image Library widget embeds an image of a structure based on PDB ID.
The Image Library widget embedding medium and small size images of the structure 4HHB.
These widgets bring RCSB PDB functionality to any resource. For more information and other widgets, see http://www.rcsb.org/pdb/static.do?p=widgets/widgetShowcase.jsp.
The structure of rat liver vault at 3.5 angstrom resolution has been split into PDB entries 2ZUO, 2ZV4, and 2ZV5. The Structure Summary page for all of these entries displays an image of the complete composite structure. Clicking on "View in Jmol" will launch a Jmol display with the composite structure.
08-December-2009
New Feature: Improved Display of Large Structures
Images and Jmol displays on the RCSB PDB website now show the complete biological assembly for all structures--even for proteins that are split across multiple PDB coordinate files.
A number of structures in the PDB archive are so large that the historical limitations of the PDB file format (maximum of 99999 atoms and 62 unique chains) require them to be split across multiple PDB coordinate files. Examples include extremely large ribosome complexes (e.g., 1GIX, 1GIY), and structures that contains a very large number of atoms or chains, such as the vault protein (e.g., 2ZUO, 2ZV4, 2ZV5).
These structures are identified on Structure Summary pages in the new "Split Entry" box, which lists and links the PDB IDs of all entries that make up the composite structure. A link is provided to easily download all of the related coordinate files.
Images on the Structure Summary page now illustrate the full composite structure, rather than what is found in each individual PDB coordinate file. The image on the Structure Summary page for 1GIX, for example, shows the full ribosome structure composed of entries 1GIX and 1GIY. For all Structure Summary images, the forward and backward buttons can be used to toggle between the asymmetric unit and the biological assembly (or multiple biological assemblies, if applicable) for the full structure.
The biological assembly and asymmetric unit of the composite structure can be launched in Jmol when viewing the corresponding static image. The display of such large structures in Jmol is possible by loading PDB files limited to CA and P atoms, and by using the Jmol load files command.
See the WHAT'S NEW page to learn about other recently-added features and improvements.
01-December-2009
Upcoming Meetings: ASCB
From December 5-9, the RCSB PDB will be at the 49th Annual Meeting of the American Society for Cell Biology in San Diego, CA. Stop by exhibition booth 730 to meet with members of the RCSB PDB and the PSI Structural Genomics Knowledgebase (PSI SGKB). David Goodsell, author of the Molecule of the Month, will discuss "Visual Representation from Atoms to Cells: New Work from the Machinery of Life" as part of a session on Exploring Cell Biology at the Frontier of 3D Visualization (Saturday, December 5, Room 15A). The PSI SGKB will be presenting a poster (Sunday, December 6, board B713) and a tutorial (Monday, December 7, 5:45pm, Room 25A) on the "One-Stop Shop For Protein Structure, Function, Sequence, Methods, and More."
24-November-2009
New Website Features: Improved Tabular Reports
For any search result, users can examine individual PDB entries or review the entire set of structures by creating reports that can be viewed online or downloaded. Recently, this reporting system has been enhanced with improved functionality and navigation, and now offers better support for reporting on very large result sets.
Report Navigation
Reports can be generated for structures matching a query by selecting any of the prepared reports available from the pulldown menu or by creating a new, customized report. Instead of presenting the entire report on a single page, reports are now available on multiple, customizable pages. By default, the first 15 records sorted by PDB ID are shown, with an option to list more records per page.
The entire table can be sorted by clicking on the column headers. All column widths are resizable by dragging the line between two columns. Within the reports themselves, PDB IDs link to that entry's Structure Summary page, PubMed IDs to the abstract, and Ligand IDs to a Ligand Summary page.
Exporting Reports
Tables can be exported in three formats: Excel 97-2003, Excel 2007 or newer, and CSV, or Comma Separated Value format (recommended for extra large data sets that may surpass size limitations in Excel).
The Excel spreadsheets are formatted with customized column width, text wrapping, alignment, and hyperlinks on selected columns.
Generating Large Reports
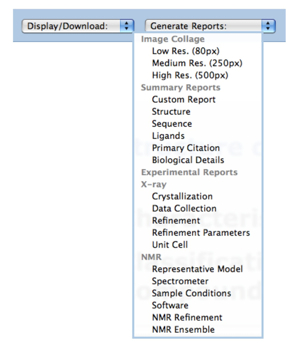
A new feature is the ability to generate reports with extremely large sets of data. For example, a report displaying the X-ray Refinement Details (Rvalue, Rfree, and Resolution) can be made for all crystal structures in the PDB.
To generate this report, select "PDB Statistics" at top right of home page, and click on the first link for "Summary Table of Released Entries".
Selecting the number for X-RAY / Total will return the more than 53000 structures that match this query. From the navigation bar, click on "Generate Reports" and select the report Experimental Reports >> X-ray >> Refinement. This report can be browsed, formatted, and downloaded as described above.
Other large reports, such as the summary reports for Biological Details and Sequence, can be easily generated for all structures in the PDB.
The latest website release also includes enhancements to browsing query results, viewing large structures, and more. See the WHAT'S NEW page for detailed descriptions.
17-November-2009
New Website Features
The latest website release includes enhancements to browsing query results, generating tabular reports, viewing large structures, and more. See the WHAT'S NEW page for detailed descriptions.
Newsletter Published
The fall 2009 issue (HTML | PDF) describes recent outreach activities and data deposition and data query news.
In the Education Corner, Gary Battle reports on a recent symposium that demonstrated the different ways the Cambridge Structural Database is used to teach chemistry. In the Community Focus, Roland Dunbrack, Jr. (Fox Chase Cancer Center) discusses his research in computational structural biology and the software his laboratory makes available for download.
For printed copies, please send your postal address to info@rcsb.org.
10-November-2009
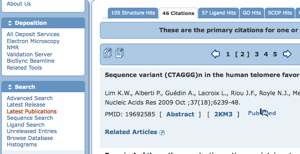
Latest Publications: How to Access All Articles in Each Update
Available from the left-hand menu, the new "Latest Publications" search returns the PubMed articles associated with PDB structures in the most recent update. This includes publications for newly released structures and for structures whose PubMed abstracts were added to our database in that update.
The results are presented in the Query Results Browser with the "Citations" tab highlighted by default. The other query results tabs are provided as well, so by clicking on the "Structure Hits" tab, all structures associated with the publications can be explored.
Clicking on the first icon in the blue bar above the citations (
 ) downloads all citations in Medline format.
) downloads all citations in Medline format.
03-November-2009
DOIs for PDB Structures and the Molecule of the Month
PDB structures can be cited using their PDB ID and the related published citation.
Structures may also be referenced using their Digital Object Identifier (DOI). These DOIs are formatted as 10.2210/pdbXXXX/pdb - where XXXX is the PDB ID. For example, the DOI for entry 4HHB is "10.2210/pdb4hhb/pdb".
This DOI can be used in a URL (http://dx.doi.org/10.2210/pdb4hhb/pdb) or entered in a DOI resolver (such as http://www.crossref.org/) to automatically link to pdb4hhb.ent.Z in the main PDB FTP archive.
DOIs are also available for each RCSB PDB Molecule of the Month feature with the format: 10.2210/rcsb_pdb/mom_YYYY_MM (where YYYY is the year, and MM the number of the month, one or two digits). For example, the DOI for the May 2003 feature on hemoglobin by Shuchismita Dutta and David S. Goodsell is "10.2210/rcsb_pdb/mom_2003_5". An RCSB PDB Molecule of the Month feature may be referenced using the DOI and the author/s of the article.
A page describing policies & references for using and citing PDB data and RCSB PDB resources is available.
27-October-2009
New and Improved Web Services for Accessing PDB Data
Web Services can help software developers build tools that interact more effectively with PDB data. Instead of storing coordinate files and related data locally, web services let software tools to interact with the RCSB PDB remotely. Documentation for accessing the RCSB PDB's Web Services is available (RESTful | SOAP).
RESTful services exchange XML files in response to URL requests. RESTful search services return a list of IDs for Advanced Search and SMILES-based queries. RESTful fetch services return data when given IDs, including PDB entity descriptions, ligand information, third-party annotations for protein chains, and PDB to UniProtKB mappings. SOAP Web Services are also available.
Improvements are being made based on community feedback. Please let us know if there are website options that you think should be offered as a web service.
20-October-2009
Poster Prize Awarded at ECM
Yonca Yuzugullu was awarded the RCSB PDB Poster Prize at the 25th European Crystallographic Meeting (August 16-21, 2009; Istanbul, Turkey) for Crystallization of bifunctional catalase-phenol oxidase (CATPO) from Scytalidium thermophilum. Yonca Yuzugullu, Chi Trinh, Arwen R. Pearson, Mark A. Smith, Simon Phillips, Ufuk Bakir, Michael J. McPherson, Zumrut B. Ogel (Middle East Technical University, Turkey).
Many thanks to Elspeth Garman (University of Oxford) and Joel Sussman (Weizmann Institute of Science) for judging the posters, and to Andreas Roodt (University of the Free State, South Africa) for organizing the judging.
13-October-2009
Nobel Prize Awarded for the Structure of the Ribosome
Three structural biologists have won the 2009 Nobel Prize in Chemistry for studies of the structure and function of the ribosome -- Venkatraman Ramakrishnan (MRC Laboratory of Molecular Biology), Thomas A. Steitz (Yale University), and Ada E. Yonath (Weizmann Institute of Science). The depositions of their first complete ribosome subunit structures (1fjg, 1ffk, and 1fka) almost a decade ago ushered structural biology into a new era. Since that time, more than 120 ribosome structures consisting of 50S, 30S subunits and complete 70S ribosomes have been contributed by these Nobel scientists. The structures, complexed with and without antibiotics, tRNAs, mRNAs, initiation factors, and release factors, provide a basis for understanding how the ribosome works and are useful tools for drug development.
The ribosome was highlighted in the October 2000 edition of the Molecule of the Month. Animations of the large and small subunits of the ribosome are available for display and download.
More information about this award is available at nobelprize.org and NIGMS.
06-October-2009
wwPDB FTP Advisory Notice
Four changes will be made to the wwPDB FTP site on Nov 24, 2009.
- The script for mirroring the FTP site using the RSYNC program will be modified to prompt the user to choose one of three rsync servers (RCSB PDB, PDBe, PDBj).
- The top-level README file will point to the download instructions hosted on the wwPDB website.
- The directory of newsletters will be updated.
- Sequence cluster data that is used only by the RCSB PDB website will be removed from the wwPDB FTP site. The data will be made available from the RCSB PDB at
ftp://resources.rcsb.org/sequence/clusters/. These data are currently at:
RCSB PDB: ftp://ftp.wwpdb.org/pub/pdb/derived_data/NR/
PDBe: ftp://ftp.ebi.ac.uk/pub/databases/rcsb/pdb-remediated/derived_data/NR/
PDBj: ftp://pdb.protein.osaka-u.ac.jp/pub/pdb/derived_data/NR/
Questions should be sent to the wwPDB at info@wwpdb.org.
29-September-2009
SF-Tool: A Tool for Crystallographic Experimental Data Validation
A streamlined, web-based tool is available for validating crystallographic experimental data. SF-Tool can be used to:
- Validate model coordinates against structure factor data (using SFCheck)1
- Easily translate your structure factor file between different formats (mmCIF, MTZ, CNS/CNX, XPLOR, SHELX, TNT, HKL2000, SCALEPACK, D*Trek, SAINT, or OTHER format)
- Check for twinned or detwinned data
Documentation for this program is available from the SF-Tools page. Questions, comments, and suggestions should be sent to deposit@deposit.rcsb.org.
1A.A. Vaguine, J. Richelle, S.J. Wodak (1999) SFCHECK: A unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D55:191-205.
22-September-2009
What's New at the RCSB PDB?
The WHAT'S NEW button at the top of the RCSB PDB header highlights recently-released tools and enhancements to the website. Descriptions, examples, and in some cases, screencasts illustrate how to access and use these features. Recent examples include new options available for reviewing query results and the Comparison Tool for calculating pairwise sequence and structure alignments.
Detailed news about all RCSB PDB activities is published online each week, and in a quarterly newsletter (click here to subscribe online or in print).
We welcome your questions and comments about these new features.
15-September-2009
New Tool For Exploring Sequence and Structure Alignments
The new RCSB PDB Comparison Tool can calculate pairwise sequence and structure alignments using a variety of methods. This feature is available on the Compare Structures web page and as a downloadable web widget.
This functionality is also integrated with the Sequence Clusters offered from each entry's Sequence Similarity tab, for example 4HHB. Users can select a pair of chains from a given sequence cluster (95% for this example), and then run either sequence or structure alignments.
The current sequence alignments possible are: blast2seq, Needleman-Wunsch, and Smith-Waterman; the structure alignments offered are FATCAT, Mammoth, TM-Align, and TopMatch. FATCAT alignments can be viewed on the external server at fatcat.burnham.org/fatcat or through the RCSB PDB's new JFatCat tool. JFatCat is a 3D structure alignment program based on FATCAT, BioJava and Jmol. JFatCat can be launched from the Comparison Tool or downloaded to the desktop.
08-September-2009
Improved Navigation of the RCSB PDB Website
RCSB PDB web pages been reorganized to make navigating the website and search results easier and more intuitive.
The left-hand menu now groups frequently-used webpages into sections -- Deposition, Home, Search, Tools, and Education -- that can be moved up and down to create a left-hand menu ordered by user interest. This customized menu will then appear on every webpage. Links that appeared in the old left-hand menu system have been moved to more contextual places, such as query result pages.
By default, search results now display the most recently-released structure first. Results can also be sorted by PDB ID, Residue Count, and Resolution.
Query result pages offer different tabs for reviewing any Structure Hits, Unreleased Structures, Citations, Ligand Hits, Web Page Hits, and GO, SCOP, and CATH Hits. Options for viewing, downloading, and generating reports for these search results appear as icons at the top of each page. Mousing over the icon indicates what action will be performed. A detailed description of the new result browser options is available at:

ACA Poster Prize Winner Magdalena Korczynska with ACA President Dr. Robert Von Dreele.
01-September-2009
Poster Prize Awarded at ACA
Magdalena Korczynska was awarded the RCSB PDB Poster Prize at the 2009 Annual Meeting of the American Crystallographic Association (ACA; July 25-30; Toronto, Canada) for Structural Insight into Homoserine Transacetylase from Haemophilus influenzae (Magdalena Korczynska (1), I. Ahmad Mirza (1), and Albert M. Berghuis (1,2); (1)Departments of Biochemistry and (2) Microbiology & Immunology, McGill University, Montreal, QC, Canada).
Many thanks to the judges: Joe Ng (The University of Alabama in Huntsville), John Rose (University of Georgia), and Emil Pai (University of Toronto).
25-August-2009
Beta Release of Redesigned BioSync

New capabilities and descriptions of operational synchrotron beamlines worldwide are now available at biosync-beta.rutgers.edu. The new site also offers search functions for services and equipment. BioSync has been redesigned to include the dramatic changes in data collection capabilities at synchrotron beamlines (including remote data collection, mail-in, crystallization and structure solution services, robotics handling for crystal screening and mounting, microfocus beams and facilities for collecting data under extreme conditions). The new look and feel of the website helps users find information about particular beamlines and to search for capabilities, services and equipment. The website's enhanced interface lets synchrotron personnel dynamically edit and add data.
BioSync offers deposition statistics by each synchrotron site and by geographical region. Galleries of structures and tables containing citations and other general information (e.g. phasing methods, resolution, R-factors, numbers of atoms) are also available. A separate set of statistical tables, galleries and informational tables is provided for structures produced by structural genomics efforts.
BioSync was formed in 1990 as a grassroots organization for the promotion of access to synchrotron radiation for scientists whose primary research was in the field of structural biology. In 1995, PDB depositions from synchrotron data came from 20 beamlines spread out over 7 different sites, and they accounted for 17% of the total number of X-ray structures deposited that year. By 2000, there were more than 60 beamlines collecting data on macromolecules and the percentage of X-ray structures collected on these beamlines was at 62%. This growth in demand was foreseen by the members of BioSync and they suggested that a web resource be created to help structural biologists identify beamlines suited to their particular experiments and the BioSync web resource went online in 2000. By 2009, the number of beamlines used to collect data on biological macromolecules has more than doubled and they account for close to 80% of current X-ray structures deposited to the PDB.
The redesign and upgrade of BioSync is being funded by NIGMS.
18-August-2009
New Opportunities to Solve and Study Protein Structures with PSI:Biology

NIGMS has released three funding opportunities that will establish the core of the PSI:Biology research network, which will apply high-throughput structural approaches to solve interesting biological problems:
- Centers for High-Throughput Structure Determination: large-scale centers that will have the capacity to solve structures on the order of several hundred per year.
- Centers for Membrane Protein Structure Determination: small centers that will devote special effort to solving the structures of these proteins.
- Consortia for High-Throughput Enabled Structural Biology Partnerships: these awards will support functional studies of proteins proposed by individuals or groups of researchers from across all fields of biology as well as support the structural determination of those proteins through consortia with the PSI:Biology structure centers. Ideal projects will integrate functional and structural data for a large number of protein structures to solve significant biological problems.
Additional program announcements for experimental technology development, computational and molecular modeling will be announced.
11-August-2009
Newsletter Published
The summer 2009 issue (HTML | PDF) highlights the new features and functionality that have been added to www.pdb.org. In the Community Focus, Gregory Warren discusses the worlds of modeling and crystallography. Recent outreach activities, along with data deposition and data query news, are also available.
For printed copies, please send your postal address to info@rcsb.org.

04-August-2009
Turn Your Computer into a PDB Structure Kiosk
Highlight structures from your lab, institution, or class with the Molecules in Motion Kiosk Viewer. Using a list of PDB IDs, this full-screen animation program will display any PDB structure from different angles and perspectives. It also focuses on any chemical components within the structure. The Java viewer can be downloaded or launched from the Educational Resources page.
28-July-2009
Poster Prize Awarded at ISMB
The RCSB PDB Poster Prize at the Joint 17th Annual International Conference for Intelligent Systems for Molecular Biology (ISMB) and the 8th European Conference on Computational Biology (ECCB; June 27-July 2; Stockholm, Sweden) went to Christoph Malisi for Automated Scaffold Selection for Enzyme Design (Christoph Malisi (1)(2), Oliver Kohlbacher (2) and Birte Höcker (1); (1) Max Planck Institute for Developmental Biology, Protein Design Group, Tübingen, Germany; (2) Division for Simulation of Biological Systems, Center for Bioinformatics, Eberhard-Karls-Universität Tübingen, Germany).
Many thanks to the judges: Marco Punta (Committee Chair, Columbia University), Yana Bromberg (Columbia University), Stefano Lise (University College London), Silvio Tosatto (Universita' di Padova, Italy), Maria Valentini (CRS4, Italy), Curtis Huttenhower (Harvard University), and Lars Arvestad (KTH Royal Institute of Technology, Sweden). Thanks also to the full reviewing committee and to the International Society for Computational Biology.
21-July-2009
Upcoming Meetings: ACA
From July 25-30, the RCSB PDB will be at the 2009 Meeting of the American Crystallographic Association in Toronto, Canada. Stop by exhibition booth 418 to meet with members of the RCSB PDB and the PSI Structural Genomics Knowledgebase (PSI SGKB). On Tuesday, July 28, the poster "The PDB: Updates and Future Plans" will describe recent improvements made to the PDB File Format and future plans for a common wwPDB deposition and annotation tool. The RCSB PDB Poster Prize will be awarded to the best student presentation related to macromolecular crystallography.
14-July-2009
Upcoming Meetings: Protein Society
At the 23rd Annual Symposium of The Protein Society (July 25-29, Boston, MA), the RCSB PDB will be exhibiting alongside the PSI Structural Genomics Knowledgebase (PSI SGKB) in booth #122. On Sunday, July 26, be sure to stop by the poster A Lot More Than Coordinates: A Short Tour of a PDB File. Presented by Lihua Tan, this poster will highlight all of the rich information available in a PDB file other than the X, Y, Z coordinates. Also on Sunday, the PSI SGKB will host a workshop that will describe How to Use the PSI Structural Genomics Knowledgebase for Research (Salon B, 12:00-1:00 p.m.).
07-July-2009
Sequence Similarity Views of PDB Structures
Interested in finding homologous protein structures, or finding a non-redundant set of proteins? The new Sequence Similarity View is an easy way to get this information based upon a protein chain in a PDB entry. This view, available through a tab at the top of each Structure Summary page, provides clusters of structures at different levels of sequence similarity. Details about the structures of all homologous proteins found within a cluster can then be examined.
The clusters are based on a weekly BLAST analysis of all proteins with more than 20 amino acids in the PDB. Many PDB entries contain several chains, so the sequence similarity is defined on a chain-by-chain basis, with the results returned for the entire structure. For more information, see the description at http://www.pdb.org/pdb/statistics/clusterStatistics.do.
To see proteins that are from 30% to 100% identical with the alpha and beta chains of hemoglobin, explore the Sequence Similarity View for 4hhb.
30-June-2009
wwPDB News: Gerard Kleywegt to head Protein Data Bank Europe
Starting July 1, 2009, Gerard Kleywegt will lead the Protein Data Bank Europe (PDBe) project at the European Bioinformatics Institute (Hinxton, UK). During the last 17 years he has been working in Uppsala, Sweden, a center of excellence for biomolecular crystallography and has developed many tools that are widely used by structural biologists worldwide. He has an extensive publication record and has served on the PDBe Scientific Advisory Board for the last few years. He has also been one of the European representatives on the wwPDB advisory committee. He has a very strong international reputation and is well respected in the structural biology community.
Gerard will be replacing Kim Henrick, who has been the team leader of the PDBe since 2001. During his tenure at the EBI, Kim has made enormous contributions to the PDBe at the EBI, by establishing and leading a strong team that has developed a wide variety of services. Kim was a strong advocate for the formation of the wwPDB in 2003. He has played a key role in developing the standards for representing all the data in the archive from small molecules to very large biological assemblies, and in leading the remediation efforts. Kim's deep knowledge of the PDB, his very critical eye, and sharp wit will be greatly missed by the wwPDB.
The members of the wwPDB look forward to working closely with Gerard and wish Kim all the best in the next chapter of his career.
23-June-2009
ISMB/ECCB and Other Upcoming Meetings
Stop by exhibit booth #9 at the Joint 17th Annual International Conference for Intelligent Systems for Molecular Biology (ISMB) and the 8th European Conference on Computational Biology (ECCB) to visit with the RCSB PDB and the PSI SGKB (June 27-July 2; Stockholm, Sweden).
Also at this meeting, the RCSB PDB Poster Prize will be awarded for the outstanding student poster in the structure and function prediction category. At the ISMB satellite Bioinformatics Open Source Conference (BOSC), the RCSB PDB's Andreas Prlić will present BioJava 2009: an Open-Source Framework for Bioinformatics, and at the BioLink Special Interest Group Session on the Future of Scientific Publishing, Phil Bourne will discuss OpenID vs. ResearcherID.
Exhibition booths and posters will also be presented at the 23rd Annual Symposium of The Protein Society (July 25-29; Boston, MA) and the 2009 Annual Meeting of the American Crystallographic Association (July 25-30; Toronto, Canada).
At the ASBMB's Essentials for Educating Biochemistry and Molecular Biology Undergraduates Symposium (August 5-8; Colorado Springs, CO), the RCSB PDB's Shuchismita Dutta will run a workshop entitled Molecular Visualization and Protein Databases (IIA): Tools, Rules and Stories: A Protein Data Bank Workshop Series.
16-June-2009
How does an HPUB structure get released?
An HPUB status indicates that a structure will be released when the primary reference is published. When it's confirmed that the corresponding article is available, the structure is included in the weekly update of the PDB.
The wwPDB receives publication dates and citation information directly from a few journals. For most articles, however, the wwPDB searches PubMed and scans the literature for publication information. Citations emailed to deposit@wwpdb.org are also greatly appreciated.
There is a one-year limit on the length of a hold period, including HPUBs. If the citation for a structure is not published within the one-year period, depositors will be given the option to either release or withdraw the deposition.
09-June-2009
Customizable Structure Summary Pages
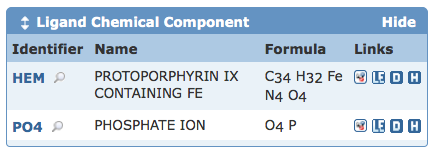
Tabbed Structure Summary pages let users explore information about any given structure. Located above the structure ID and title, each tab offers different types of information -- a general overview, derived data, sequence details, sequence similarity, literature view, biology & chemistry, methods, geometry, and links to external resources. Data are organized on each page in different blocks, or "widgets". On a structure's Summary and Derived Data pages, these widgets can be hidden or expanded, and in some cases, moved around on the page. Users can organize these pages to highlight information of interest.
An online video demonstrates how to customize these page layouts
Customized layouts are automatically stored in a cookie and are persistent for all PDB IDs. To switch back to the default layout, users should click on the "Reset View" in the left-hand menu.
This enhancement to Structure Summary pages is is one of many new features added to the RCSB PDB website -- a full list is available.
02-June-2009
Literature View: Looking at Structures in PubMedCentral
The recently-released Literature View aims to provide a broad look at how a given structure has been analyzed and presented in open access publications, that is, where the full text of the article is available without copyright restriction. The overall intent is to make the PDB user more aware of publications associated with the structure under study. It is one of many new features added to the RCSB PDB website -- a full list is available.
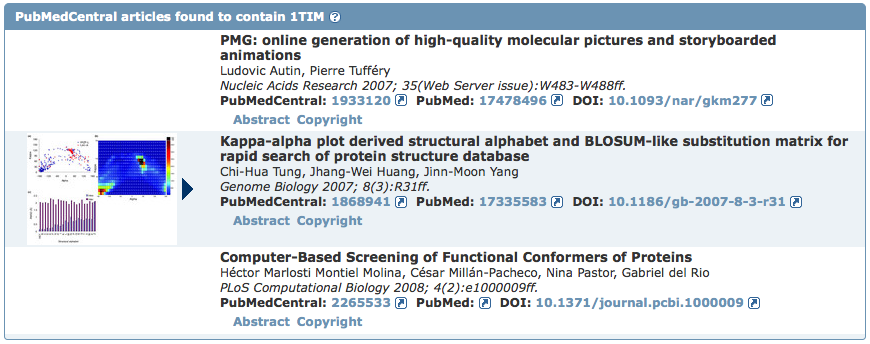
Accessible by selecting the Literature tab from any Structure Summary page, this view highlights a variety of published articles associated with a PDB entry. The primary citation's abstract is included and can be used to search for structures with PubMed abstracts containing the same keywords. The Literature View also indicates the other publications included in the entry by the authors in addition to the primary citation (those contained in REMARK 1).
In collaboration with the BioLit project (biolit.ucsd.edu), the Literature View lists any open access articles found in PubMedCentral that contain the entry's PDB ID including papers that don't include the PDB ID's primary citation in the references. Links to the abstract and copyright information, along with figures and related legends from these articles are displayed.
If the listed open access articles also mention other PDB entries, their image, PDB ID, title, and sequence similarity to the initial structure are included in a table with links to their Structure Summary pages.
To see some examples, explore the Literature View for entries 1TIM, 1O4X, 2G12, and 1AOI.
26-May-2009
Release of new design, features, and functionality
The modernized look of the RCSB PDB website compliments the new features and functionality released this week. A full "What's New" list for this release is available, with some of the new features highlighted below:
|
Quick searches by PDB ID/keyword, author name, structural genomics center, and chemical component name and ID are available from a new pulldown menu offered on every page. |

|
|
Customizable Structure Summary pages let users highlight particular types of information. Boxes ("widgets") can be clicked and dragged, and their content hidden or shown. New layouts are automatically stored for future visits. A video screencast demonstrates how to customize the layout of this page.
These pages use icons to indicate links to help pages (?), molecular viewers ( |

|
|
The Ligand Explorer Viewer available on the Structure Summary page can be used to dynamically view hydrogen bond, hydrophobic, bridged H-bond, metal interaction, and neighbor residue interactions and thresholds. |

|
|
Other new features include a Literature Tab View that incorporates PubMedCentral abstracts and images, and a Sequence Similarity Tab View that provides provides an overview of the sequence clusters for each protein chain in a PDB structure. |

|
We look forward to any comments about these new features!
19-May-2009
5 Easy Steps for Structure Deposition
Depositors can follow these steps to make the process of depositing a structure quick, easy, and accurate!
- Check the sequence, which should contain all residues used in the experiment, including expression tags and residues missing due to disorder. Use BLAST1 to determine the appropriate sequence database references for the proteins or nucleic acids present in the file. Any sequence mismatches should correspond to mutation, variant or expression tags in the submitted sequence.
- Check the ligands. Ligand Expo can be used to see if any of the chemical components in the structure (ligands, drugs, inhibitors, ions, modified residues, etc.) already exist in the Chemical Component Dictionary. If found, use the corresponding 3-character code in the coordinates; otherwise, the new component can be uploaded in ADIT along with the structure.
- Prepare data for deposition. Use pdb_extract to automatically collect information needed for deposition from the output files produced by many structure determination applications, and SF-Tool to convert the structure factor file format and check the data with SFCHECK.2
- Validate the structure with the Validation Suite and Server to ensure that the data being deposited are accurate and reflect what you intend to submit. This program checks the sequence and file format consistency, compares geometrical and chemical interactions to various standards, and reports errors that should be corrected before deposition.
- Deposit using ADIT/ADIT-NMR to check, validate, and edit the PDB data entries.
Deposition is an iterative process. If you encounter problems at a particular step, please make the correction(s) and go through the steps again.
These steps, along with a checklist of items needed for deposition, are described in an updated brochure called 5 Easy Steps for Structure Deposition. For a printed copy or for a packet of information about the deposition process, please send your postal address to info@rcsb.org.
1S.F. Altschul, W. Gish, W. Miller, E.W. Myers, & D.J. Lipman (1990) Basic local alignment search tool. J. Mol. Biol. 215:403-410.
2A.A. Vaguine, J. Richelle, S.J. Wodak (1999) SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D55:191-205.
12-May-2009
Looking at Structures: A Resource for Learning About PDB Data
Where are all the hydrogen atoms in this file? Should I care about the R-factor? Why are there 20 overlapped structures in my file? These questions and many others are explored in the RCSB PDB's new Looking at Structures.
Using text, images, and interactive Jmols, Looking at Structures intends to help researchers and educators get the most out of the PDB archive. Broad topics include how to understand PDB data, how to visualize structures, how to read coordinate files, and potential challenges in exploring the archive.
A Table of Contents appears on the right side of every page so at any time users can access the individual pages: Biological Units, Dealing with Coordinates, Methods for Determining Structure, Missing Coordinates and Biological Units, Molecular Graphics Programs, Resolution, and R-value and R-free.
Looking at Structures is available from the General Education section of the left-hand menu.
05-May-2009
Ligand Expo: Searching and Browsing Features
Ligand Expo can be used to access chemical and structural information about all small molecule components found in PDB entries.
The latest release includes amino acid variants--protonation and de-protonation states--as part of Ligand Expo's Browse option. For example, when browsing arginine, protonation states ARG_LEO2 and ARG_LFOH_RNH1 will be included in the results list.
Ligand Expo can also Search for ligands using:
- files in a variety of formats including PDB, MOL/SDF and the Refmac/Phenix monomer library (mmCIF)
- chemical name
- formula
- SMILES STRING
- 3-character ID code
Ligand Expo is based upon the Chemical Component Dictionary maintained by the wwPDB. Ligand Expo's capabilities have been described online and in a flyer available for download.
28-April-2009
MyPDB: Keep up-to-date with new structures...automatically!
MyPDB notifies users via email when the PDB releases structures that match customized queries. With MyPDB, users can:
- combine and save keyword, sequence, ligand, and other searches
- run searches stored in MyPDB at any time
- receive email alerts weekly or monthly
- access structures directly from the email alerts for further exploration
- utilize the RCSB PDB web interface to refine saved searches
Information on how to register and use MyPDB is available in the Spring RCSB PDB Newsletter.
21-April-2009
Improve the Quality of Your Depositions with SFCheck
SFCheck is integrated with the RCSB PDB's deposition tools so users can assess the agreement between an atomic model and the corresponding X-ray data before a structure is deposited and released.
SFCheck generates reports containing R-factors, density correlation, Luzzati plots, Wilson plots, temperature factors, local error estimation by residues, and more. It is a part of the Validation Server and ADIT, and is used by the RCSB PDB during the annotation process.
SFCheck is also part of SF-Tool, a streamlined, web-based tool for validating crystallographic experimental data. In addition to validating model coordinates against structure factor data with SFCheck, SF-Tool can:
- Easily translate a structure factor file between different formats (mmCIF, CIF, MTZ, CNS/CNX, XPLOR, SHELX, TNT, HKL2000, SCALEPACK, XSCALE, D*Trek, SAINT, other)
- Check for twinned or detwinned data
Depositors are strongly encouraged to use SFCheck before deposition.
SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model AA Vaguine, J Richelle, SJ Wodak (1999);
Acta Crystallogr D Biol Crystallogr 55:191-205.
14-April-2009
Newsletter Published
In the spring 2009 issue (HTML | PDF), David Goodsell interviews molecular animators Gaël McGill and Graham Johnson to find out how PDB data gets translated into their detailed movies and images. In the Education Corner, Gavin Whittaker describes how physical models can be created and used to provide a unique view of molecular structure. New RCSB PDB features and developments, including the recent release of MyPDB email alerts, are highlighted.
For printed copies, please send your postal address to info@rcsb.org.
Exhibition at Experimental Biology
Please stop by the RCSB PDB's exhibition booth at the 2009 Experimental Biology meeting (April 18-22) in New Orleans, Louisiana. Staff will be available at booth #1410 to answer questions and to demonstrate the many deposition and query features available from www.pdb.org.
07-April-2009

Crystallography for Modelers: May 7-8, 2009
The RCSB PDB will host a short course for practicing pharmaceutical/biophysical modelers looking for a better understanding of crystal structures and PDB data. The course will be held May 7 & 8 at Rutgers, The State University of New Jersey in Piscataway, NJ.
Do you use protein crystal structure data? Ever wonder why things do not work out as you expect? Are ligand strain energies unreasonably high? Does your drug candidate not appear to make the hydrogen bonds you expect? Is that really a water molecule in the corner of a binding site? Do you wonder why a side chain is in one position rather than another?
These questions all relate to the "quality" of the data you use. How "good" are the protein structures that you use? What's the error in atomic coordinates? How might you know if something is just plain wrong?
Crystallography for Modelers will address these issues and offer insight into state-of-the-art research into the quality, errors, and "gotchas" in crystal structure data. Designed by active users of the data, the instructors are experts from RCSB PDB team members, Rutgers researchers, and commercial software developers. It is not an introductory course for crystallography or modeling, but rather a down-and-dirty discussion of crystallographic data, precision, accuracy, and possible errors.
Registration is for Day 1, Day 2 or both. Day 2 will expand on Day 1 and offer an opportunity for hands-on learning.
Day 1: The process of crystal structure determination. Insight into the workings of the PDB and how "PDB" files are generated. Detailed examination of the extensive information (beyond coordinates) available in data files and online, including ligand structures. Proper interpretation of crystal structures, part one: accuracy, precision, problems and errors (experimental, real, perceived).
Day 2: Proper interpretation of crystal structures, part two: case studies with emphasis on ligand binding, position, conformation, loops, and missing density. Software bridging modeling and crystallography. Hands-on exercises in the concepts of the course with public and commercial software. Software vendor showcase.
To learn more and register: rate.rutgers.edu/?q=Crystallography
Both classes will take place at Rutgers' Busch Campus in Piscataway, NJ, which is easily accessible by car, train, or air.
31-March-2009
Bridgewater-Raritan High School Wins New Jersey Science Olympiad Protein Modeling State Finals
|
After their win at the Central Regional tournament, the team from Bridgewater-Raritan High School came in first place at the protein modeling event at the 2009 New Jersey Science Olympiad.  (Click image to enlarge) The models were carefully judged by RCSB PDB staff.  (Click image to enlarge) Livingston High School. In an extremely close competition, West Windsor-Plainsboro High School South came in second, and Livingston High School came in third out of 24 schools. Teams from all over the Garden State submitted their hand-built 3D models of a ribonuclease protein, along with an abstract, to be judged by staff from the RCSB PDB. At the competition, teams built a model of a selected region of a ribonuclease structure using Jmol and completed a written exam. Teams used the Molecule of the Month and other RCSB PDB resources, to help prepare for this event. Congratulations to all participating teams--there were many great models, abstracts, and responses to the written exam. The final results, pictures, and a video demonstrating how models are built for this competition are available at education.pdb.org. Questions about the NJ Science Olympiad Protein Modeling trial event should be sent to buildmodels@deposit.rcsb.org. Special thanks to our judges from the RCSB PDB (Shuchismita Dutta, Chenghua Shao, Hyumni Sun, Christine Zardecki (Event Supervisor) and Marina Zhuravleva ), the NJ Science Olympiad organizers, and to the MSOE Center for BioMolecular Modeling for the design of this event. |
24-March-2009

RCSB PDB at the San Diego Science Festival April 4
Come build virus models with the RCSB PDB at Expo Day on April 4th in Balboa Park, San Diego, CA.
As part of the San Diego Science Festival (SDSF), Expo Day will offer a full day of hands-on activities for every type of science-enthusiast. In addition to building viruses, scientists will be on hand at the RCSB PDB booth at the Expo to help visitors explore the many structures found in the PDB archive. Visitors will also learn the latest ways to conserve water resources and discover how the cars of tomorrow will mulch vegetable oil into fuel. Interactive activities, comedians and stage shows will captivate and excite any aspiring scientist.
SDSF is planned as one of the nation's largest multi-cultural, multi-generational, multi-disciplinary celebrations of science. It will bring together students, families, businesses, scientists, and communities for a festival that is interactive and fun, highlighting the impact of science and innovation on our lives. The Science Festival is hosting FREE activities throughout March and the beginning of April. On April 1, high school students from all over San Diego County will visit UCSD for an invited event that will include tours of PDB structures in UCSD's CAVE environment. Supported by private and corporate sponsors, local colleges and universities, leading research institutes, and hundreds of community and science organizations, SDSF showcases what is truly unique about science in San Diego.
50 Years of Protein Structure DeterminationThe NIGMS has documented 50 years of advances in high-resolution protein structure determination in an interactive timeline. Beginning in 1958 with the publication of the first structure in atomic detail, the timeline charts more than 25 advances over five decades. The entries include brief descriptions, images and citations. |
17-March-2009
PDB Archive Version 3.15 Released
A newly standardized and enhanced version of the entire PDB archive at ftp://ftp.wwpdb.org has been released.
Files originally released before December 2, 2008 will follow PDB Format Version 3.15; files originally released after that date will follow Version 3.20. For detailed file format documentation, please see www.wwpdb.org/docs.html.
A time-stamped snapshot of the PDB archive before this release is available from https://snapshots.wwpdb.org/ in the directory 20090316.
Users who maintain local copies of the wwPDB FTP will have to download the entire archive. Scripts to help in this process are available at www.wwpdb.org/downloads.html.
These data reflect the wwPDB's continuing commitment to providing accurate and detailed data to users worldwide. This release includes improvements and enhancements to the data, including details about the chemistry of the polymer and the ligands bound to it, biological assemblies, and binding sites of ligands and metal ions. An overview (PDF) is provided at the wwPDB website.
Questions may be sent to info@wwpdb.org.
Receive Email Alerts When New Structures Match Your Queries with MyPDB
MyPDB is a new feature that regularly sends out emails when structures that match customized queries are released. The matching structures can be accessed directly from the email alerts. Users can also log in to MyPDB to run stored searches at any time.
To sign up for MyPDB, users should register using the link at the top right of the RCSB PDB header.

Following registration, an activation link is sent in a confirmation email. After activating the MyPDB account, users can log in at the top of the page.

Next, users can review currently saved queries and update MyPDB account information. This page can also be accessed by clicking on the user name at the top of the page.

Saved MyPDB Queries can be keyword, sequence, ligand, and Advanced Searches. To store a query in a profile, users should query the RCSB PDB website, such as an Advanced Search for structures with Enzyme Commission Number 2.7.11. After evaluating the query, the "Queries" tab in the left menu will temporarily store the search. Selecting "Save Query to MyPDB" option from this tab will store the search in MyPDB.

From the MyPDB page, users can click on the query name and query description to personalize them.

By default, the email notification is turned on. The "Next Scheduled Run" indicates when the query is next run. Email alerts can be set to run weekly (Tuesday afternoon Pacific Time) or monthly (first Tuesday each month).
The email includes the customized query name and a list of PDB IDs and titles matching the query. Clicking on the query name takes users to the results set at the RCSB PDB website; the results can then be further refined, exported, or further explored. Each PDB ID links to the entry's Structure Summary page.
Please let us know if you have any questions about this new feature.
10-March-2009

Crystallography for Modelers: May 7-8, 2009
The RCSB PDB will host a short course for practicing pharmaceutical/biophysical modelers looking for a better understanding of crystal structures and PDB data. The course will be held May 7 & 8 at Rutgers, The State University of New Jersey in Piscataway, NJ.
Do you use protein crystal structure data? Ever wonder why things do not work out as you expect? Are ligand strain energies unreasonably high? Does your drug candidate not appear to make the hydrogen bonds you expect? Is that really a water molecule in the corner of a binding site? Do you wonder why a side chain is in one position rather than another?
These questions all relate to the "quality" of the data you use. How "good" are the protein structures that you use? What's the error in atomic coordinates? How might you know if something is just plain wrong?
Crystallography for Modelers will address these issues and offer insight into state-of-the-art research into the quality, errors, and "gotchas" in crystal structure data. Designed by active users of the data, the instructors are experts from RCSB PDB team members, Rutgers researchers, and commercial software developers. It is not an introductory course for crystallography or modeling, but rather a down-and-dirty discussion of crystallographic data, precision, accuracy, and possible errors.
Registration is for Day 1, Day 2 or both. Day 2 will expand on Day 1 and offer an opportunity for hands-on learning.
Day 1: The process of crystal structure determination. Insight into the workings of the PDB and how "PDB" files are generated. Detailed examination of the extensive information (beyond coordinates) available in data files and online, including ligand structures. Proper interpretation of crystal structures, part one: accuracy, precision, problems and errors (experimental, real, perceived).
Day 2: Proper interpretation of crystal structures, part two: case studies with emphasis on ligand binding, position, conformation, loops, and missing density. Software bridging modeling and crystallography. Hands-on exercises in the concepts of the course with public and commercial software. Software vendor showcase.
To learn more and register: rate.rutgers.edu/?q=Crystallography
Both classes will take place at Rutgers' Busch Campus in Piscataway, NJ, which is easily accessible by car, train, or air.
PDB Archive Version 3.15 to be Released March 17, 2009*
The wwPDB will release a newly standardized and enhanced version of the entire PDB archive at ftp://ftp.wwpdb.org with the March 17, 2009 update. *Please note the revised date.
Files originally released before December 2, 2008 will follow PDB Format Version 3.15; files originally released after that date will follow Version 3.20. For detailed file format documentation, please see www.wwpdb.org/docs.html.
A time-stamped snapshot of the PDB archive before this release will be available from https://snapshots.wwpdb.org/.
Users who maintain local copies of the wwPDB FTP will have to download the entire archive. Scripts to help in this process are available at www.wwpdb.org/downloads.html.
These data reflect the wwPDB's continuing commitment to providing accurate and detailed data to users worldwide.
Questions may be sent to info@wwpdb.org.
03-March-2009
PSI:Biology Phase Approved
In January 2009, the NIGMS Council approved the PSI's next 5-year plan of research, to be called PSI:Biology. This new phase will concentrate on applying the technologies developed in the PSI Pilot and Production phases to study a broader range of biological and biomedically relevant scientific problems. More information can be found in the official NIGMS news release and at the PSI SG Knowledgebase.
24-February-2009Exhibition at the Biophysical Society MeetingThe RCSB PDB will participate in the exhibition at the 53rd Annual Meeting of the Biophysical Society (February 28 - March 4) in Boston, Massachusetts. Staff will be available at booth #603 to answer questions and to demonstrate the many deposition and query features available from www.pdb.org. We hope to see you there! |
17-February-2009
PDB Archive Version 3.15 to be Released March 2009
The wwPDB will release a newly standardized and enhanced version of the entire PDB archive at ftp://ftp.wwpdb.org in March 2009.
Files originally released before December 2, 2008 will follow PDB Format Version 3.15; files originally released after that date will follow Version 3.20. For detailed file format documentation, please see www.wwpdb.org/docs.html.
A time-stamped snapshot of the PDB archive before this release will be available from https://snapshots.wwpdb.org/.
Users who maintain local copies of the wwPDB FTP will have to download the entire archive. Scripts to help in this process are available at www.wwpdb.org/downloads.html.
These data reflect the wwPDB's continuing commitment to providing accurate and detailed data to users worldwide.
Questions may be sent to info@wwpdb.org.
10-February-2009
Available only as a download, the poster
(31Mb) "Molecular Machinery: A Tour of the Protein Data Bank" features illustrations of 75 select structures from the PDB, showing their relative sizes at a scale of three million to one, and generally describes their critical roles in the functions of living cells. A 2-sided, 8 1/2" x 11" quick reference guide (5Mb) is also available.
Tools for Education
The General Education section of the RCSB PDB offers a variety of resources for teachers and students interested in learning about protein and nucleic acid structures.
Accessible from the left hand menu, this section offers:
- Looking at Structures: Through text, images, and interactive Jmols, this feature intends to help researchers and educators get the most out of the PDB archive. Broad topics include how to understand PDB data, how to visualize structures, how to read coordinate files, and potential challenges in exploring the archive.
- Educational Resources: This page archives many of the educational materials created by the RCSB PDB--animations, instructional handouts, and much more. Recently, lectures, lesson plans, and related activities about viruses for middle and high school classrooms have been added. This page also catalogues events such as the New Jersey Science Olympiad and the RCSB PDB Poster Prize.
- RCSB PDB Newsletters and Education Corner: Published quarterly, the newsletter describes new developments, outreach efforts, and interviews with leaders in the community. The newsletter's Education Corner, which highlights how the PDB is used in different facets of education, are also archived on a single page. Past Education Corners have featured the program Molecules for the iPhone, fruit-flavored protein folding, and creating molecular multimedia in the college classroom.
- Proteins in the Sea: An interactive kiosk at the The Sea of Genes exhibit at the Birch Aquarium (La Jolla, CA) displayed information about specific proteins found in marine organisms--and in the PDB. The Flash animation used in the exhibit and a flyer (PDF) describing it are available
- Molecule of the Month: Each Molecule of the Month installment includes an introduction to the structure and function of a particular molecule, a discussion of the relevance of the molecule to human health and welfare, suggested additional readings and resources, and interactive views.
Other resources are described in a downloadable flyer (PDF).
03-February-2009
wwPDB Paper: Deposition and Annotation
wwPDB deposition tools, methods (including validation), and policies have been described in:
Molecular Biotechnology (2008) [
doi:10.1107/S0907444908017393 ]
Data Deposition and Annotation at the Worldwide Protein Data Bank
Shuchismita Dutta, Kyle Burkhardt, Jasmine Young, Ganesh J. Swaminathan, Takanori Matsuura, Kim Henrick, Haruki Nakamura and Helen M. Berman.
A list of all related RCSB PDB publications is available from the RCSB PDB website. Reprints may be requested by contacting info@rcsb.org.
27-January-2009
NJ Science Olympiad Protein Modeling Results
Many ribonuclease models were built by high school students for the RCSB PDB-sponsored
Protein Modeling event at the New Jersey Science Olympiad Regionals.
| Northern Regional 1 Westfield 2 Livingston 3 Montville |
Central Regional 1 Bridgewater-Raritan HS 2 West Windsor-Plainsboro HS South 3 Princeton HS |
Southern Regional 1 MATES Academy 2 Moorestown Friends School 3 Eastern Regional HS |

(Click image to enlarge) Bridgewater-Raritan High School.

(Click image to enlarge) A judge described how the models were scored with the team from Westfield.

(Click image to enlarge) Annotators judged the protein models.

(Click image to enlarge) The MATES Academy team built a portion of ribonuclease at the event.
Before the day of competition, teams build a model of the full ribonuclease structure based on PDB ID 1rta. In this model, teams are encouraged to include "additions" that help tell the functional story of ribonuclease. They also provide an abstract/key that describes features highlighted in their model.
At the event itself, teams have 50 minutes to build a section of an assigned PDB structure and answer questions about the structure, function, importance, and history of ribonuclease. For all sections of the event, students are encouraged to use the Molecule of the Month, the PDB file, Jmol and the Structure Explorer page.
2009 regional results, pictures, and tips can be found at education.pdb.org/olympiad.
Special thanks to our judges from the RCSB PDB (Shuchismita Dutta, Margaret Gabanyi, Irina Persikova, Monica Sekharan, Chengua Shao, Hyumni Sun, Lihua Tan, Huangwang Yang, Jasmine Young, Christine Zardecki, and Marina Zhuravleva ), the NJ Science Olympiad organizers, and to the MSOE Center for BioMolecular Modeling for the design of this event. We look forward to seeing everyone at the state finals in March!
20-January-2009
Time-stamped Copies of PDB Archive Available via FTP
A time-stamped snapshot of the PDB archive (ftp.wwpdb.org) as of January 5, 2009 has been added to ftp://snapshots.rcsb.org/. Snapshots of the PDB have been archived annually since 2004. It is hoped that these snapshots will provide readily identifiable data sets for research on the PDB archive.
The script at ftp://snapshots.rcsb.org/rsyncSnapshots.sh may be used to make a local copy of a snapshot or sections of the snapshot. The directory 20090105 includes the�55,072�experimentally-determined coordinate files that were current at that time. Coordinate data are available in PDB, mmCIF, and XML formats. The date and time stamp of each file indicates the last time the file was modified.
2009 NIGMS Workshop: Enabling Technologies for Structural Biology
The PSI and NIGMS have announced that the annual workshop 2009 NIGMS Workshop - Enabling Technologies for Structural Biology will be held March 4-6, 2009 at the NIH Natcher Conference Center in Bethesda, MD. This workshop will focus on the technical challenges of protein structure determination and present the latest technological and methodological innovations.
Many issues in structural genomics and structural biology are as applicable to the individual bench scientist as they are to high-throughput research centers. The preliminary program includes keynote presentations by Michael Rossmann and Frank Raushel, six topical sessions, and a poster session. All classes of protein from uncharacterized hypothetical proteins to membrane proteins and protein-protein complexes will be discussed.
For further information regarding the 2009 NIGMS Workshop, online registration, and abstract submission, visit www.blsmeetings.net/2009workshop_enablingtechnologies. The deadline for submitting an abstract is February 11, 2009, and selected abstracts will be invited to present their results.
13-January-2009
Newsletter Published
The winter 2009 edition offers tips for depositing multiple related structures and describes the release of PDB Archive Version 3.15. Deposition and website statistics for 2008, the domain annotations in 3D view, the release of the 2008 Annual Report, and the publication of a new resource for learning about PDB Data, called Looking at Structures, are described.
In this quarter's Education Corner, Bernadette Uzzi talks about protein structure-related courses available from Brookhaven National Laboratory's Science Learning Center. In the Community Focus, Johannes Kirchmair and Gerhard Wolber (University of Innsbruck) discuss their paper The Protein Data Bank (PDB), Its Related Services and Software Tools as Key Components for In Silico Guided Drug Discovery, and more.
For printed copies, please send your postal address to info@rcsb.org.
06-January-2009
Getting Started with the RCSB PDB Website
Not sure how to find what you're looking for on this website? The Getting Started page has been updated to help users access all of the data and related resources available from the RCSB PDB. This introduction offers a Quick Start to using the website and explains the tabbed navigation system and left hand menus.
For example, selecting each tab offers rich ways of exploring individual structures and search result sets. The left-hand menus organize resources by topic. Teachers, for example, will be interested in the General Education section of the website, while researchers depositing structures should look at the Deposit and Validate resources.
Other introductory resources include a narrated tutorial that illustrates how to search, navigate, browse, generate reports, and visualize structures. The Molecule of the Month also provides a good introduction to the types of structures available in the PDB archive. And as always, please let our help desk know if you have any questions