Welcome
RCSB Protein Data Bank (RCSB PDB) enables breakthroughs in science and education by providing access and tools for exploration, visualization, and analysis of:
Experimentally-determined 3D structures from the Protein Data Bank (PDB) archive | |
Computed Structure Models (CSM) from AlphaFold DB and ModelArchive |
These data can be explored in context of external annotations providing a structural view of biology.
Latest Entries
Features & Highlights
Paper Published: ZMPY3D
Read about Accelerating protein structure volume analysis through vectorized 3D Zernike moments and Python-based GPU integration
Paper Published: Using Augmented Reality
Read about Using augmented reality in molecular case studies to enhance biomolecular structure-function explorations in undergraduate classrooms
Deprecation of FTP File Download Protocol on November 1, 2024
wwPDB to deprecate FTP download protocol in the PDB archive
Poster Prize Awarded at LACA
The wwPDB Foundation awarded Zaighum Abbas at the Latin American Crystallographic Association Meeting for The Role of SEPT7 in Magnaporthe oryzae: Structural Insights and Functional Implications
The Nobel Prize in Chemistry 2024
Congratulations to David Baker, who was recognized for computational protein design, and to Demis Hassabis and John M. Jumper, who were recognized for protein structure prediction. These achievements also celebrate the work of the PDB depositor community and the PDB archive that underpinned development of their prediction methods.
Register for the November 4 Mol* Virtual Office Hour
Join RCSB PDB for quick tips on how to use Mol* to view Sequence Annotations in 3D
wwPDB Data Release Policy on Public Preprint Archives
Contributions to public preprint archives that mention PDB/EMDB/BMRB entry IDs are deemed by the wwPDB to be publications
Register for the November 7 Virtual Office Hour on Supporting Extended PDB IDs
Interested in learning more about plans to extend PDB IDs to 12 characters? Bring you questions to a virtual Office Hour.
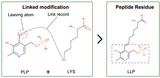
Annotation of Protein Modifications in the PDB
Protein chemical modifications (PCMs) and post translational modifications (PTMs) annotation is now included in CCD files, and updated atomic coordinate files are being rolled out.
Register Now for Webinar: Unlock Rapid Analyses Across the Whole PDB Using BinaryCIF
Join us on November 4 to future-proof your data analysis with BinaryCIF, a fully interchangeable yet drastically more efficient flavor of the PDBx/mmCIF format
See new feature archive
Education Corner: Paper Nucleic Acid Models for Hands-on Education
Cassandra Hayne and Phoebe Rice (The University of Chicago) describe downloadable Paper Nucleic Acid Models for Hands-on Education» 11/25/2024Paper Published: Impact of the PDB on drug approvals
Open access to the PDB facilitated discovery/development of 100% of the 34 new low molecular weight, protein-targeted, antineoplastic agents approved by the US FDA 2019–2023.» 11/19/2024Meet RCSB PDB at ABRCMS UPDATE!!!!!!!
Visit the poster Streamlining Programmatic Access to Structural Biology Data with Python to learn about recent software development; meet Rutgers members in booth #1740 to learn about the summer internships and gap year opportunities» 11/12/2024Paper Published: IHMCIF
Read about An Extension of the PDBx/mmCIF Data Standard for Integrative Structure Determination Methods» 11/05/2024Meet RCSB PDB at SACNAS
Learn about summer internship opportunities and more at the Rutgers booth #648» 10/27/2024Interview with Director Emerita Helen Berman
Read about The huge protein database that spawned AlphaFold and biology's AI revolution in Nature» 10/20/2024Happy Birthday, Irving Geis
Celebrate Geis' birthday (October 18, 1908) with a tour of the Geis Digital Archive of his pioneering works of biomolecular art at PDB-101» 10/17/2024PDB-101 Focus: Peak Performance
PDB-101 materials explore the structural biology of athletics and well-being. Learn how the ten enzymes of glycolysis break down sugar in our diet in Glycolytic Enzymes» 10/15/2024Paper Published: MolViewSpec Toolkit
Learn about Describing and Sharing Molecular Visualizations Using the MolViewSpec Toolkit» 10/10/2024
Molecule of the Month

Angiotensin and Blood Pressure
Many medications for controlling high blood pressure inhibit the action of the peptide hormone angiotensin.
Read MoreQuarterly News (see archive)
Issue 102 - June 2024
New Training Opportunities; A Biocurator Milestone; Building 3D Paper Models of Protein Domains; and more.
Cell signaling through Rube Goldberg Machines by Dr. Fatahiya Kashif, Federal Medical College, Islamabad, Pakistan
Annual Reports

2023 Annual Report
Download the 2023 Annual Report (PDF) for an overview of recent activities, including RCSB PDB APIs and pairwise alignment tools.