Structural basis for synthase activation and cellulose modification in the E. coli Type II Bcs secretion system.
Anso, I., Zouhir, S., Sana, T.G., Krasteva, P.V.(2024) Nat Commun
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
(2024) Nat Commun
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cellulose synthase catalytic subunit [UDP-forming] | 908 | Escherichia coli | Mutation(s): 0 Gene Names: bcsA, yhjO, yhjP, b3533, JW5665 EC: 2.4.1.12 | 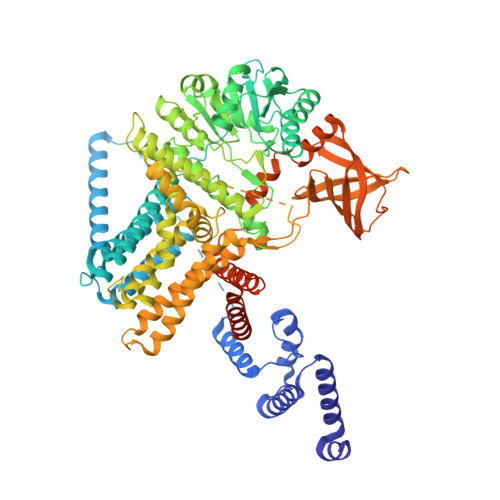 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cyclic di-GMP-binding protein | B, K [auth X], L [auth Y], M [auth Z] | 779 | Escherichia coli | Mutation(s): 0 Gene Names: bcsB, A8C65_00280, AC789_1c39010, ACU57_05345, AM464_10380, BHS81_21120, BK292_24560, BON72_13325, BON75_10020, BON76_21165... | 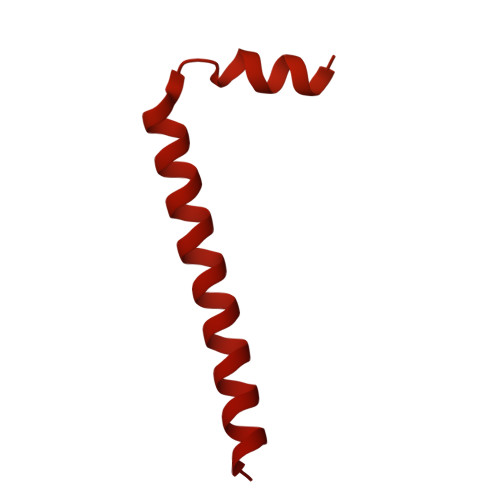 |
UniProt | |||||
Find proteins for A0A061KLG7 (Escherichia coli) Explore A0A061KLG7 Go to UniProtKB: A0A061KLG7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A061KLG7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cyclic di-GMP binding protein BcsE | C [auth E], H [auth U] | 536 | Escherichia coli | Mutation(s): 0 Gene Names: bcsE, yhjS, b3536, JW3504 | 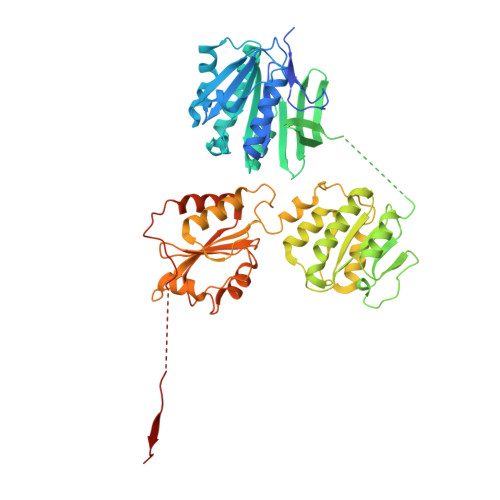 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cellulose biosynthesis protein BcsF | D [auth F], G [auth S] | 63 | Escherichia coli | Mutation(s): 0 Gene Names: bcsF, Z4953, ECs4417 | 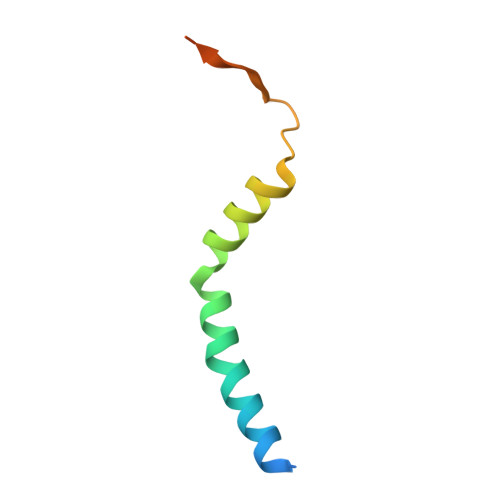 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cell division protein | E [auth Q], J [auth W] | 250 | Escherichia coli | Mutation(s): 0 Gene Names: bcsQ, yhjQ, A8C65_00290, ACU57_05335, BMT91_17060, BON75_10030, BvCmsHHP019_01723, BvCmsSINP011_05061, C2U48_15650, D3O91_11725... |  |
UniProt | |||||
Find proteins for A0A0B1KWQ0 (Escherichia coli) Explore A0A0B1KWQ0 Go to UniProtKB: A0A0B1KWQ0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0B1KWQ0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein YhjR | F [auth R], I [auth V] | 77 | Escherichia coli | Mutation(s): 0 Gene Names: yhjR, b3535, JW3503 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cellulose biosynthesis protein BcsG | N [auth G], O [auth D] | 536 | Escherichia coli | Mutation(s): 0 Gene Names: bcsG, yhjU, b3538, JW3506 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| C2E (Subject of Investigation/LOI) Query on C2E | P [auth A] Q [auth A] R [auth E] S [auth E] V [auth U] | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one) C20 H24 N10 O14 P2 PKFDLKSEZWEFGL-MHARETSRSA-N |  | ||
| ATP (Subject of Investigation/LOI) Query on ATP | T [auth Q], X [auth W] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | U [auth Q], Y [auth W] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | v4 |
| MODEL REFINEMENT | Coot | |
| MODEL REFINEMENT | PHENIX |
| Funding Organization | Location | Grant Number |
|---|---|---|
| European Research Council (ERC) | European Union | 757507 |