RCSB PDB Help
Search and Browse > Advanced Search
Overview: Advanced Search
Video: Advanced Search and Grouping
Introduction
What is Advanced Search?
Besides the 3D coordinates of atoms in the structure, all experimental structures (PDB entries) and computed structure models (CSMs) include a variety of meta-data about the experiment, polymer sequences, and ligands present in the structure. Information and annotations from other data resources are also connected to each PDB entry.
Why use Advanced Search?
RCSB PDB Advanced Search options allow you to query all data in the coordinate files and their associated annotations to rapidly find structures, polymers, and ligands relevant to the topic of interest.
Advanced Search Query Builder Options Infographic
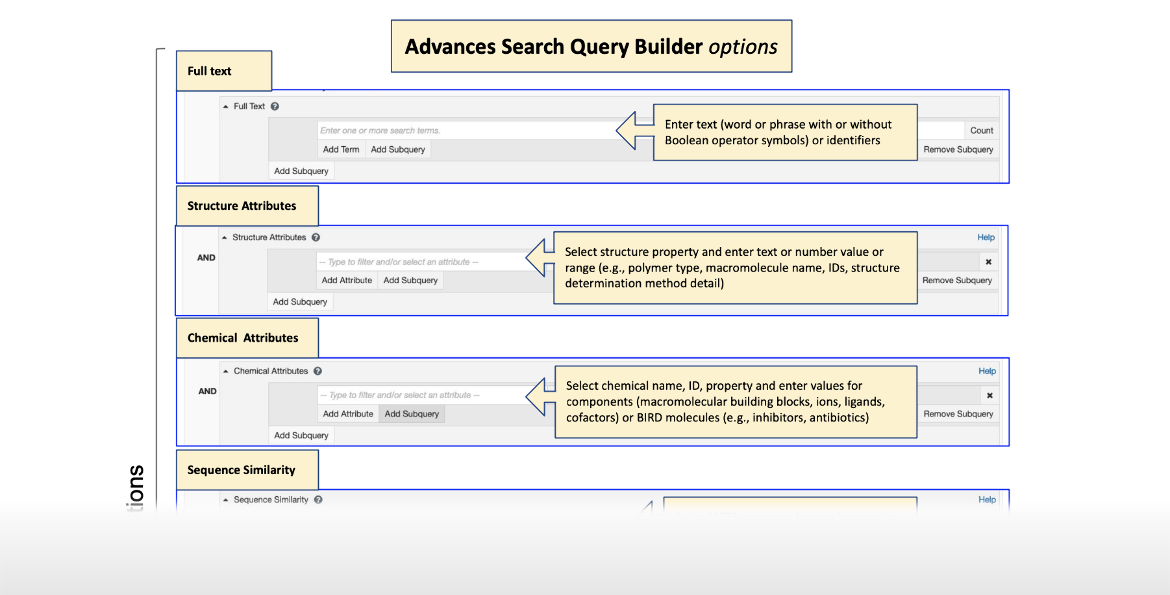
|
| Click on the image for more information ... |
Documentation
Advanced Search Query Builder Interface
The Advanced Search Query Builder interface features powerful options for constructing complex searches and managing results. Attribute queries can be seamlessly combined with sequence and structure similarity searches.
The interface also allows users to exclude or include CSMs in the search by using the default options or checking on the toggle switch next to the Count, Clear, and Search buttons (Figure 1).

|
| Figure 1: Advanced Search Query Builder options. Toggle switch on to include CSMs is shown in a red outlined box. |
Queries: Using the Advanced Search Options
The Advanced Search options allow you to construct complex composite queries to quickly find structures and/or information related to the topic of interest by
- defining the query either as specifically or as broadly as is appropriate for the search being performed.
- including or excluding CSMs in the search, and
- organizing search results in a manner that facilitates rapid identification of the individual structure matches or groups of search results of interest.
Criteria for Searching
The Advanced Search options allow you to query the archive in four distinct ways (Figure 2). For all searches, there is an option to include CSMs (by turning on the toggle switch, cyan colored) or exclude them (by turning off the toggle switch, gray colored).

|
| Figure 2: Types of Advanced Search options available from the Query builder. The option to include or exclude CSMs is also available via the cyan colored toggle switch. |
- Attribute Search: This option allows for three types of searches based on specific text-based or numerical properties of entries, assemblies, or ligands.
- Full Text searches of PDB entries and their associated annotations
- Structural Attributes searches of textual and numerical properties of PDB structure(s), their associated experimental details that relate to polymer molecules (e.g., name, identifiers)
- Chemical Attributes searches of the names and classifications of small molecules (ligands), inhibitors, drugs, etc. that are present in PDB structures
- Sequence Based Search: This option allows for searches based on the polymer sequences present in PDB structures.
- Sequence Similarity Search uses the polymer sequences of all or a significant portion of the proteins and nucleic acids present in a structure.
- Sequence Motif search uses a short polymer query sequence.
- Structure Based Search: This option enables searches based on 3D structural alignments.
- Structure Similarity Search is based on 3D shape.
- Structure Motif search is based on the local arrangement of a selected set of structural building blocks in a given PDB structure.
- Chemical Similarity Search: This option allows for searches based on chemical information (e.g., chemical formula and descriptors like SMILES, InChI).
Combining Searches
Composite and complex Boolean queries can be constructed using the 'AND', 'OR', and 'NOT' options available on the Advanced Search interface. These operators can be applied to specific attributes or groups of attributes and used as follows:
- AND: identify structures, polymers, ligands, or assemblies that meet all the specified criteria.
- OR: identify structures, polymers, ligands, or assemblies that meet any one of the specified criteria.
- NOT: exclude structures, polymers, ligands, or assemblies that meet the specified criteria.
Different search types (i.e., attribute-based, sequence-based, structure-based, and chemical information-based searches) may be combined to construct complex composite queries. As you assemble a composite query, you have the option of running subqueries to assess how many structures, polymers, etc. match those selected portions of the overall query.
Search Result Return Options
Before you launch a search there are two decisions that should be made - (a) what type of results do you wish to see; and (b) how should the results be presented (as a list of as groups)
3D structure Similarity searches can return two kinds of results:
- Structural data: PDB entry, entity, and/or assembly information pertaining to one or more structures in the archive.
- Molecular definitions: chemical component dictionary and BIRD molecule matches to specific search criteria.
Select options from the Return menu in the lower left corner of the Advanced Search Query Builder (Figure 3) about whether you wish to find 3D structures or entries (default), polymer entities (e.g., sequence matches), assembly of biological macromolecules, or small molecule ligands.
Several options for the types of search results are available in the pulldown, with the default option being “Structures”.
- Structures - PDB entries, designated with a 4-character alphanumeric identifier, e.g. 1Q2W. See grouping options for Structures.
- Polymer Entities - distinct (chemically unique) polymeric molecules present in PDB entries, specifically proteins (polypeptides), DNA (polydeoxyribonucleotide), and RNA (polyribonucleotide). See grouping options for Structures.
- Non-polymer Entities - small chemicals (enzyme cofactors, ligands, ions, etc) defined as non-polymers in the coordinate file. A non-polymer Entity ID is a combination of a PDB ID and entity ID, e.g., 4HHB_3. This option can be useful when searching for small chemicals within their macromolecular context, for instance covalently bound ligands such as ATP in 2CCH (entity 2CCH_4).
- Assemblies - the macromolecular quaternary structure believed to be the functional form of the molecule (also referred to as the “biological unit”)
- Molecular Definitions - includes standard and modified amino acids (e.g., ALA) and nucleotides (e.g., A), small molecule ligands (e.g., ATP, or HEM) as they are defined in the wwPDB Chemical Component Dictionary (CCD), and peptide-like molecules as they are defined in the Biologically Interesting molecule Reference Dictionary (BIRD), (e.g., PRD_000010).
The Results Page
Depending on the Return options selected, the search results page displays lists of 3D structures, polymer entities, ligands, assemblies etc. In all cases the results page has a Refinements menu on the left and options to view, download and create reports on the top of the page (see Figure 4).

|
| Figure 4: Part of the Search Results page showing a list of structures with icons indicating if they are experimental structures or CSMs. |
- View - the search results can be viewed in a variety of formats. In each of these views the experimental structures are marked with a dark blue colored icon showing a flask, while CSMs are marked with a cyan colored icon showing a computer. Structure identifiers of search hits shown in these views link to the respective structure summary pages. The search results may be viewed in the following formats -
- Summary - every search hit is displayed with an image and summary information.
- Gallery - every search hit is displayed with an image only.
- Compact - every search hit is displayed with summary information only.
- Tabular Report options organize results in a multi-column, multi-row format, with each row corresponding to a search hit. A set of preconfigured tabular reports is available along with options to build custom tabular reports.
- The Download Files button in the top right corner of the Search Results page allows export of the associated files for either all or a manually selected subset of search hits. Structures and experimental data can be exported in legacy PDB and mmCIF formats. Molecular definitions can be exported in .sdf and .mol2 file formats.
- The Sort by options allow you to organize the order of the returned results on the page. By default, results are sorted by relevancy scoring but can be sorted by other criteria, e.g., by release date, using the pull-down menu above the search results list. Other types of searches enabled at RCSB PDB —i.e., sequence similarity, sequence motif, structure motif, structure similarity, and chemical similarity—are powered and scored by separate strategies specific to each case. Additionally, when multiple types of searches are combined into a single search (e.g., a text + sequence similarity search), either a "combined" sorting strategy may be used (default) or users may select one of the search types to prioritize (e.g., either text or sequence similarity). More information about Sorting Strategies is available along with suggestions for how to use them to meet your needs. Besides these scoring options, search results can also be sorted by other criteria, e.g., by release date, using the pull-down menu above the search results list.
- When a large number of results are returned, the highest-ranking 25 search hits are displayed on the first page by default. To change the number of results per page click the "Page # of #" menu above the search results list to select a different number.
- The Refinements panel on the left side of the page features categories relevant to the selected "Return" as option. Checking on boxes next to relevant options and clicking on the green arrow next to the column header can For instance, if the return type is "Structure", the refinements include categories relevant to macromolecular data, e.g., taxonomy, resolution, experimental method, etc. When the "Molecular Definitions" option is selected, the refinements include categories relevant to chemical reference data, e.g., molecular weight, atom count, component type, etc.
- The Results returned are context specific and are based on the user query and the selected "Return" options. The provenance of the structure is marked with specific icons indicating if it is an experimental structure (marked with a dark blue flask icon) or a CSM (marked with a cyan colored computer terminal). Queries that return structures are listed as in Figure 4, while results of a sequence based queries displays "Polymer Entities", and the search results are enriched with pairwise sequence alignments between the sequence used in the query and the sequences of every entity returned by the search and details about the alignment (sequence identity, E value etc., see Figure 5). Learn more about sequence searches.

|
| Figure 5: Search result when the polymer return type is selected, showing sequence alignment and related details. |
Options for Grouping Results
Redundancy occurs at many levels (such as the level of sequence or structure similarity), and a variety of different grouping methods can be applied to PDB data in order to provide a non-redundant view. Available grouping options can be selected from the “grouped by” dropdown at the bottom of the Advanced Search Query Builder. Learn more about grouping search results here.















