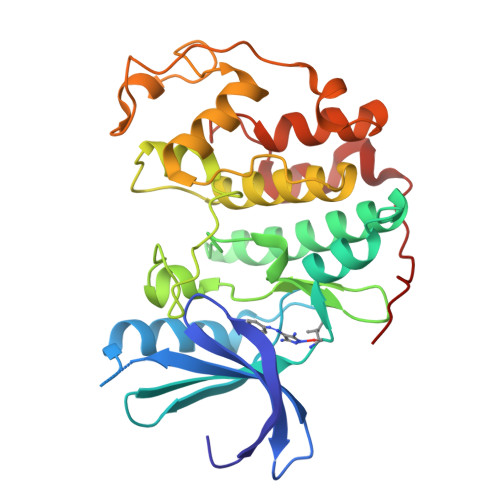
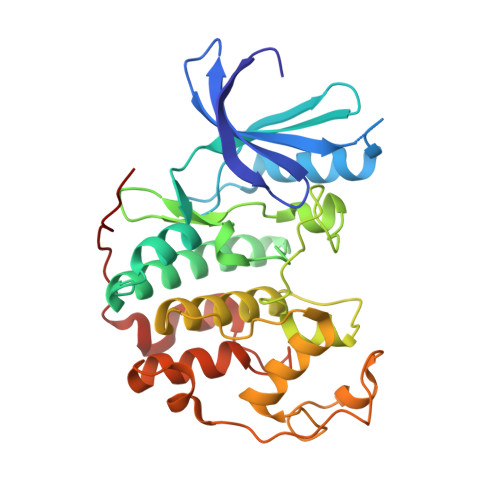
4-Arylazo-3,5-Diamino-1H-Pyrazole Cdk Inhibitors: Sar Study, Crystal Structure in Complex with Cdk2, Selectivity, and Cellular Effects
Krystof, V., Cankar, P., Frysova, I., Slouka, J., Kontopidis, G., Dzubak, P., Hajduch, M., De Azevedo Junior, W.F., Paprskarova, M., Orsag, M., Rolcik, J., Latr, A., Fischer, P.M., Strnad, M.(2006) J Med Chem 49: 6500
- PubMed: 17064068
- DOI: https://doi.org/10.1021/jm0605740
- Primary Citation of Related Structures:
2CLX - PubMed Abstract:
In a routine screening of our small-molecule compound collection we recently identified 4-arylazo-3,5-diamino-1H-pyrazoles as a novel group of ATP antagonists with moderate potency against CDK2-cyclin E. A preliminary SAR study based on 35 analogues suggests ways in which the pharmacophore could be further optimized, for example, via substitutions in the 4-aryl ring. Enzyme kinetics studies with the lead compound and X-ray crystallography of an inhibitor-CDK2 complex demonstrated that its mode of inhibition is competitive. Functional kinase assays confirmed the selectivity toward CDKs, with a preference for CDK9-cyclin T1. The most potent inhibitor, 4-[(3,5-diamino-1H-pyrazol-4-yl)diazenyl]phenol 31b (CAN508), reduced the frequency of S-phase cells of the cancer cell line HT-29 in antiproliferation assays. Further observed cellular effects included decreased phosphorylation of the retinoblastoma protein and the C-terminal domain of RNA polymerase II, inhibition of mRNA synthesis, and induction of the tumor suppressor protein p53, all of which are consistent with inhibition of CDK9.
Organizational Affiliation:
Laboratory of Growth Regulators, Faculty of Science, Palacký University and Institute of Experimental Botany, Slechtitelů 11, 783 71 Olomouc, Czech Republic.