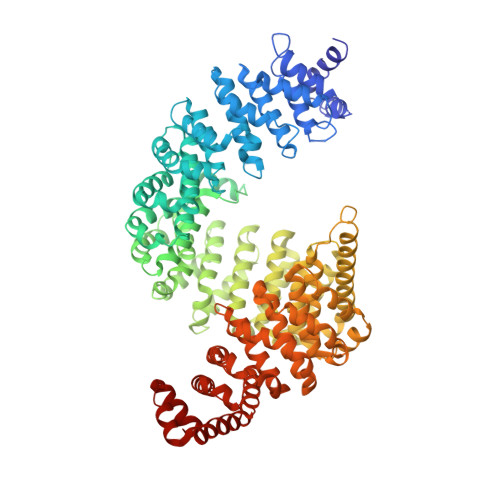
Rules for nuclear localization sequence recognition by karyopherin beta 2.
Lee, B.J., Cansizoglu, A.E., Louis, T.H., Zhang, Z., Chook, Y.M.(2006) Cell 126: 543-558
- PubMed: 16901787
- DOI: https://doi.org/10.1016/j.cell.2006.05.049
- Primary Citation of Related Structures:
2H4M - PubMed Abstract:
Karyopherinbeta (Kapbeta) proteins bind nuclear localization and export signals (NLSs and NESs) to mediate nucleocytoplasmic trafficking, a process regulated by Ran GTPase through its nucleotide cycle. Diversity and complexity of signals recognized by Kap betas have prevented prediction of new Kap beta substrates. The structure of Kap beta 2 (also known as Transportin) bound to one of its substrates, the NLS of hnRNP A1, that we report here explains the mechanism of substrate displacement by Ran GTPase. Further analyses reveal three rules for NLS recognition by Kap beta 2: NLSs are structurally disordered in free substrates, have overall basic character, and possess a central hydrophobic or basic motif followed by a C-terminal R/H/KX(2-5)PY consensus sequence. We demonstrate the predictive nature of these rules by identifying NLSs in seven previously known Kap beta 2 substrates and uncovering 81 new candidate substrates, confirming five experimentally. These studies define and validate a new NLS that could not be predicted by primary sequence analysis alone.
- Department of Pharmacology, University of Texas Southwestern Medical Center at Dallas, 6001 Forest Park, Dallas, TX 75390, USA.
Organizational Affiliation: