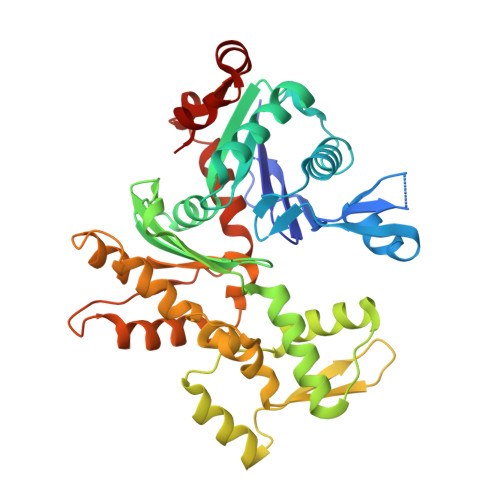
Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation.
Ducka, A.M., Joel, P., Popowicz, G.M., Trybus, K.M., Schleicher, M., Noegel, A.A., Huber, R., Holak, T.A., Sitar, T.(2010) Proc Natl Acad Sci U S A 107: 11757-11762
- PubMed: 20538977
- DOI: https://doi.org/10.1073/pnas.1005347107
- Primary Citation of Related Structures:
3MMV, 3MN5, 3MN6, 3MN7, 3MN9 - PubMed Abstract:
Three classes of proteins are known to nucleate new filaments: the Arp2/3 complex, formins, and the third group of proteins that contain ca. 25 amino acid long actin-binding Wiskott-Aldrich syndrome protein homology 2 domains, called the WH2 repeats. Crystal structures of the complexes between the actin-binding WH2 repeats of the Spire protein and actin were determined for the Spire single WH2 domain D, the double (SpirCD), triple (SpirBCD), quadruple (SpirABCD) domains, and an artificial Spire WH2 construct comprising three identical D repeats (SpirDDD). SpirCD represents the minimal functional core of Spire that can nucleate actin filaments. Packing in the crystals of the actin complexes with SpirCD, SpirBCD, SpirABCD, and SpirDDD shows the presence of two types of assemblies, "side-to-side" and "straight-longitudinal," which can serve as actin filament nuclei. The principal feature of these structures is their loose, open conformations, in which the sides of actins that normally constitute the inner interface core of a filament are flipped inside out. These Spire structures are distant from those seen in the filamentous nuclei of Arp2/3, formins, and in the F-actin filament.
Organizational Affiliation:
Max-Planck-Institut für Biochemie, 82152 Martinsried, Germany.