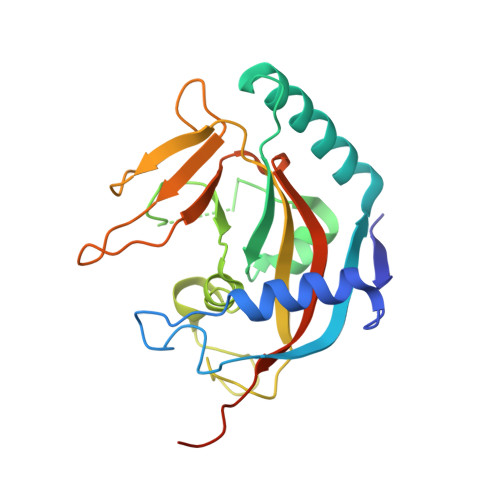
Structure of human tankyrase 1 in complex with small-molecule inhibitors PJ34 and XAV939.
Kirby, C.A., Cheung, A., Fazal, A., Shultz, M.D., Stams, T.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 115-118
- PubMed: 22297980
- DOI: https://doi.org/10.1107/S1744309111051219
- Primary Citation of Related Structures:
3UH2, 3UH4 - PubMed Abstract:
The crystal structures of tankyrase 1 (TNKS1) in complex with two small-molecule inhibitors, PJ34 and XAV939, both at 2.0 Å resolution, are reported. The structure of TNKS1 in complex with PJ34 reveals two molecules of PJ34 bound in the NAD(+) donor pocket. One molecule is in the nicotinamide portion of the pocket, as previously observed in other PARP structures, while the second molecule is bound in the adenosine portion of the pocket. Additionally, unlike the unliganded crystallization system, the TNKS1-PJ34 crystallization system has the NAD(+) donor site accessible to bulk solvent in the crystal, which allows displacement soaking. The TNKS1-PJ34 crystallization system was used to determine the structure of TNKS1 in complex with XAV939. These structures provide a basis for the start of a structure-based drug-design campaign for TNKS1.
Organizational Affiliation:
Novartis Institute for Biomedical Research, Cambridge, MA 02139, USA.