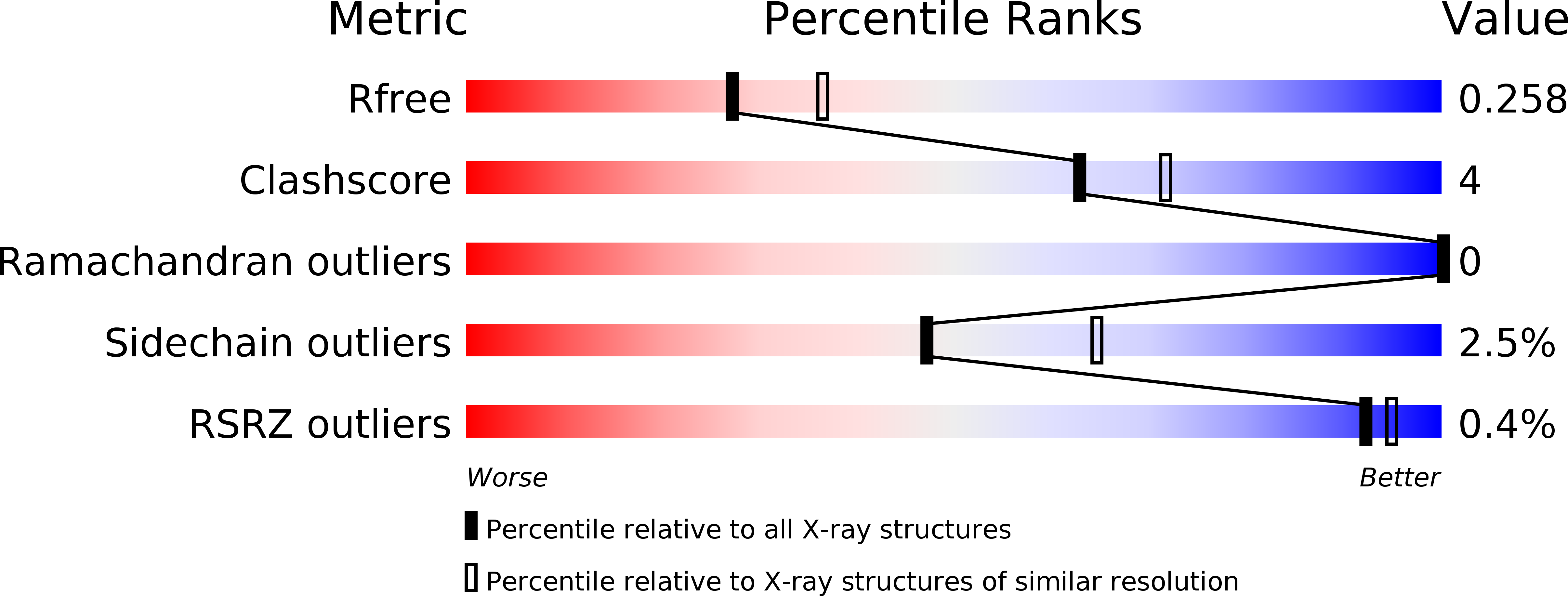
NXL104 irreversibly inhibits the {beta}-lactamase from Mycobacterium tuberculosis.
Xu, H., Hazra, S., Blanchard, J.S.(2012) Biochemistry 51: 4551-4557
- PubMed: 22587688
- DOI: https://doi.org/10.1021/bi300508r
- Primary Citation of Related Structures:
4DF6 - PubMed Abstract:
NXL104 is a novel β-lactamase inhibitor with a non-lactam structural scaffold. Our kinetic and mass spectrometric analysis demonstrates that NXL104 quantitatively inhibits BlaC, the only chromosomally encoded β-lactamase from Mycobacterium tuberculosis, by forming a carbamyl adduct with the enzyme. The inhibition efficiency (k(2)/K) of NXL104 was shown to be more than 100-fold lower than that of clavulanate, a classical β-lactamase inhibitor, which is probably caused by the bulky rings of NXL104. However, the decarbamylation rate constant (k(3)) was determined to be close to zero. The BlaC-NXL104 adduct remained stable for at least 48 h, while the hydrolysis of the BlaC-clavulanate adduct was observed after 2 days. The three-dimensional crystal structure of the BlaC--NXL104 carbamyl adduct was determined at a resolution of 2.3 Å. Interestingly, the sulfate group of NXL104 occupies the position of a phosphate ion in the structure of the BlaC-clavulanate adduct and is hydrogen bonded to residues Ser128, Thr237, and Thr239. Favorable interactions are also seen in the electrostatic potential map. We propose that these additional interactions, as well as the intrinsic stability of the carbamyl linkage, contribute to the extraordinary stability of the BlaC-NXL104 adduct.
Organizational Affiliation:
Department of Biochemistry, Albert Einstein College of Medicine, Bronx, New York 10461, United States.