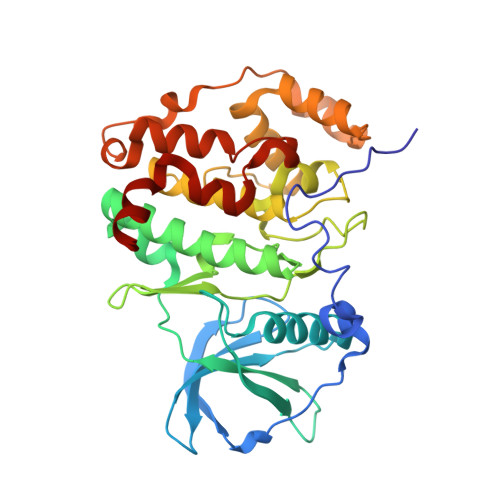
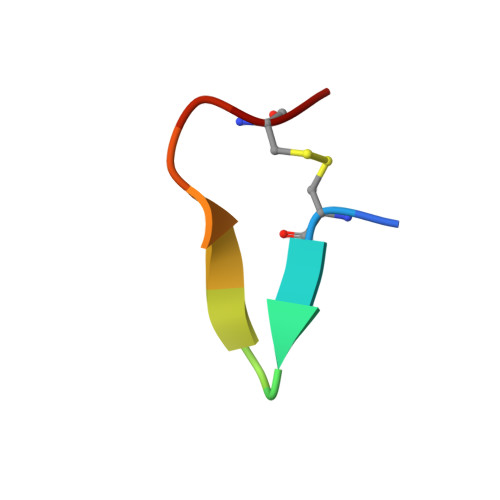
First structure of protein kinase CK2 catalytic subunit with an effective CK2 beta-competitive ligand
Raaf, J., Guerra, B., Neundorf, I., Bopp, B., Issinger, O.G., Jose, J., Pietsch, M., Niefind, K.(2013) ACS Chem Biol 8: 901-907
- PubMed: 23474121
- DOI: https://doi.org/10.1021/cb3007133
- Primary Citation of Related Structures:
4IB5 - PubMed Abstract:
The constitutively active Ser/Thr kinase CK2 (casein kinase 2) is used by tumor cells to acquire apoptosis resistance. CK2 exists as a heterotetrameric holoenzyme with two catalytic chains (CK2α) attached to a dimer of noncatalytic subunits (CK2β). A druggable cavity at the CK2β interface of CK2α allows the design of small molecules disturbing the CK2α/CK2β interaction and thus affecting activity, stability, and substrate specificity. We describe here the first structure of CK2α with an effective CK2β-competitive compound, namely, a 13-meric cyclic peptide derived from the C-terminal CK2β segment. Some well-ordered water molecules not visible in CK2 holoenzyme structures were detected at the interface. Driven mainly by enthalpy, the peptide binds with submicromolar affinity to CK2α, stimulates its catalytic activity, and reduces effectively the CK2α/CK2β affinity. The results provide a thermodynamic and structural rationalization of the peptide's CK2β-competitive functionality and pave thus the way to a peptidomimetic drug addressing the CK2α/CK2β interaction.