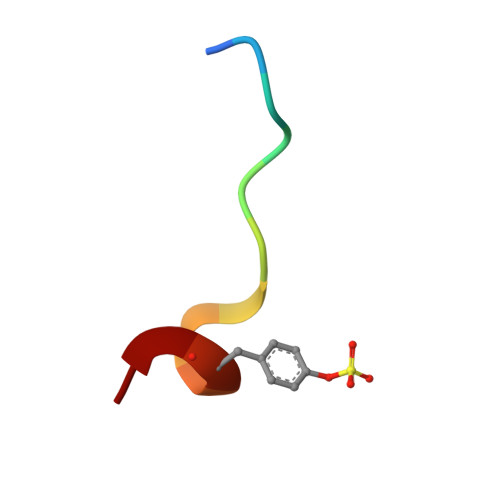
Fragments Can Bind Either More Enthalpy or Entropy-Driven: Crystal Structures and Residual Hydration Pattern Suggest Why.
Ruehmann, E., Betz, M., Heine, A., Klebe, G.(2015) J Med Chem 58: 6960
- PubMed: 26270568
- DOI: https://doi.org/10.1021/acs.jmedchem.5b00812
- Primary Citation of Related Structures:
4UD9, 4UDW, 4UE7, 4UEH, 5AF9, 5AFY, 5AFZ, 5AHG - PubMed Abstract:
In lead optimization, small, enthalpically advantaged fragments have been suggested to be superior, as an entropic component will be added inevitably during late-stage optimization. Determination of thermodynamic signatures of weak-binding fragments is essential to support the decision-making process, to decide which fragment to take to further optimization. High-resolution crystal structures of six fragments binding to the S1 pocket of thrombin were determined and analyzed with respect to their thermodynamic profile. The two most potent fragments exhibiting an amidine-type scaffold are not the most enthalpic binders; instead a chloro-thiophene fragment binds more enthalpically. Two chemically very similar chloro-aromatic fragments differ strongly in their potency (430 μM vs 10 mM); their binding modes are related, but the surrounding residual water network differs. The more potent one recruits a water molecule and involves Glu192 in binding, thus succeeding in firmly capping the S1 pocket. Fragments exhibiting a rather perfect solvation pattern in their binding mode also experience the highest potency.
Organizational Affiliation:
Institute of Pharmaceutical Chemistry, Philipps-University Marburg , Marbacher Weg 6, 35037 Marburg, Germany.