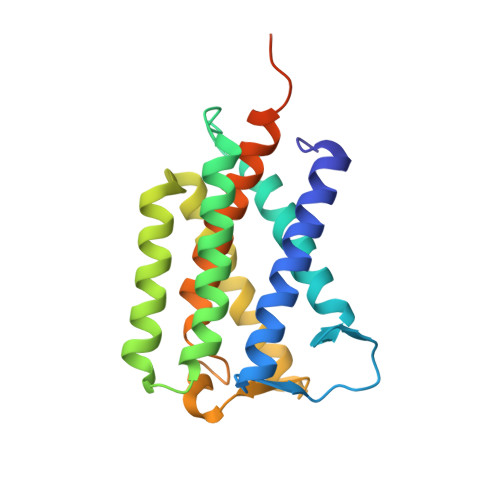
PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Ren, R., Zhou, X., He, Y., Ke, M., Wu, J., Liu, X., Yan, C., Wu, Y., Gong, X., Lei, X., Yan, S.F., Radhakrishnan, A., Yan, N.(2015) Science 349: 187-191
- PubMed: 26160948
- DOI: https://doi.org/10.1126/science.aab1091
- Primary Citation of Related Structures:
4XU4, 4XU5, 4XU6 - PubMed Abstract:
Insulin-induced gene 1 (Insig-1) and Insig-2 are endoplasmic reticulum membrane-embedded sterol sensors that regulate the cellular accumulation of sterols. Despite their physiological importance, the structural information on Insigs remains limited. Here we report the high-resolution structures of MvINS, an Insig homolog from Mycobacterium vanbaalenii. MvINS exists as a homotrimer. Each protomer comprises six transmembrane segments (TMs), with TM3 and TM4 contributing to homotrimerization. The six TMs enclose a V-shaped cavity that can accommodate a diacylglycerol molecule. A homology-based structural model of human Insig-2, together with biochemical characterizations, suggest that the central cavity of Insig-2 accommodates 25-hydroxycholesterol, whereas TM3 and TM4 engage in Scap binding. These analyses provide an important framework for further functional and mechanistic understanding of Insig proteins and the sterol regulatory element-binding protein pathway.
- State Key Laboratory of Membrane Biology, Tsinghua University, Beijing 100084, China. Center for Structural Biology, School of Life Sciences, School of Medicine, Tsinghua University, Beijing 100084, China. Tsinghua-Peking Center for Life Sciences, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: