Twist and turn: a revised structural view on the unpaired bubble of class II CPD photolyase in complex with damaged DNA.
Maestre-Reyna, M., Yamamoto, J., Huang, W.C., Tsai, M.D., Essen, L.O., Bessho, Y.(2018) IUCrJ 5: 608-618
- PubMed: 30224964
- DOI: https://doi.org/10.1107/S205225251800996X
- Primary Citation of Related Structures:
5ZCW - PubMed Abstract:
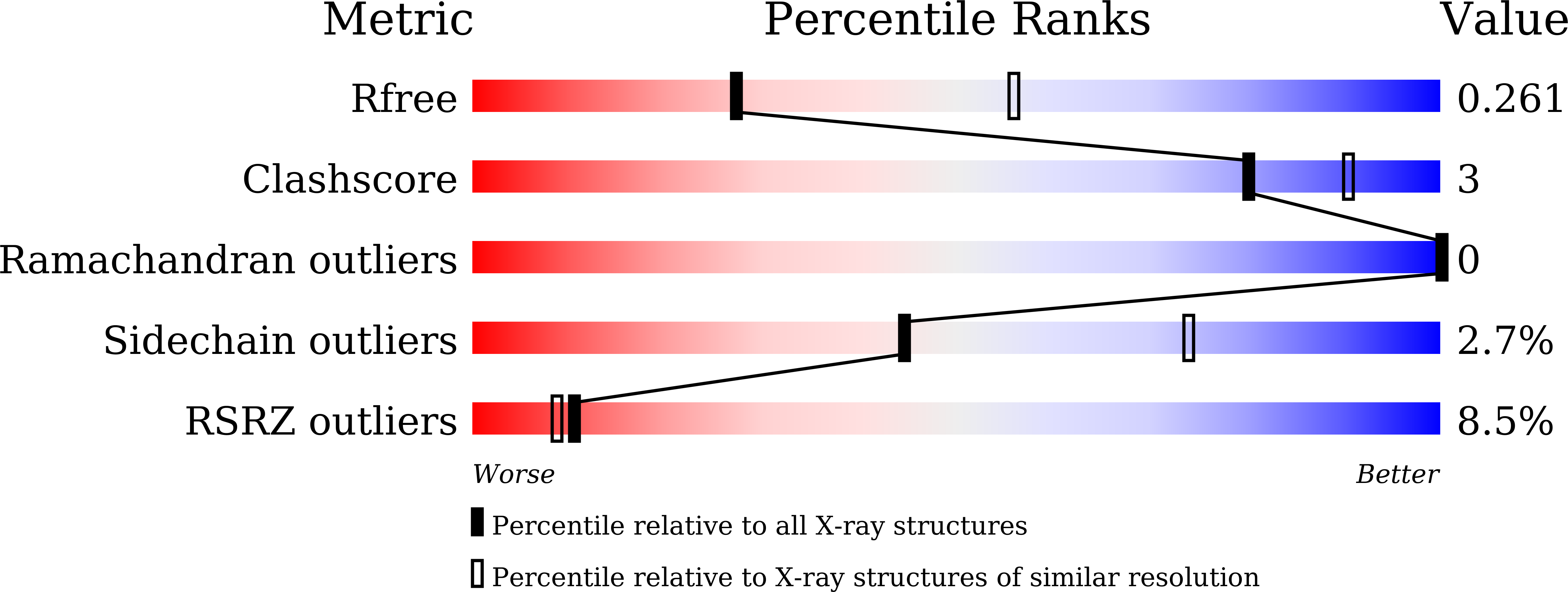
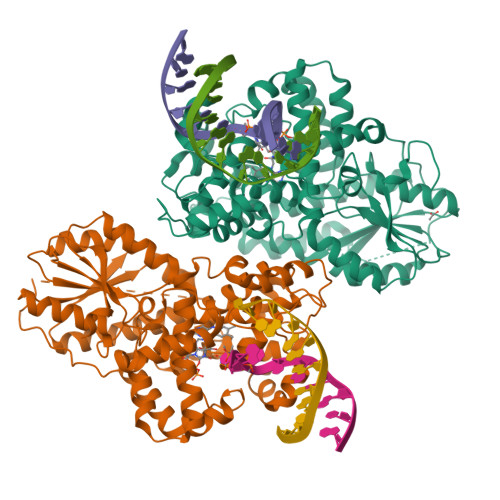
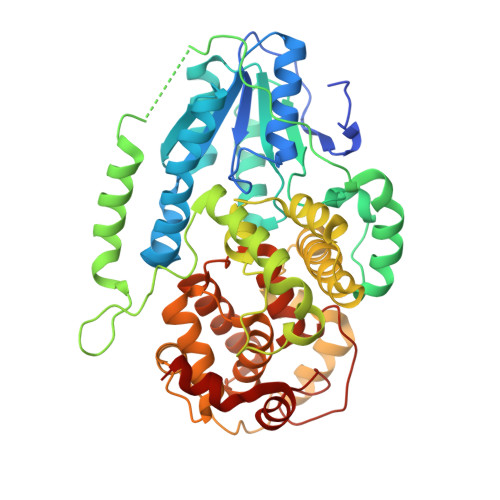
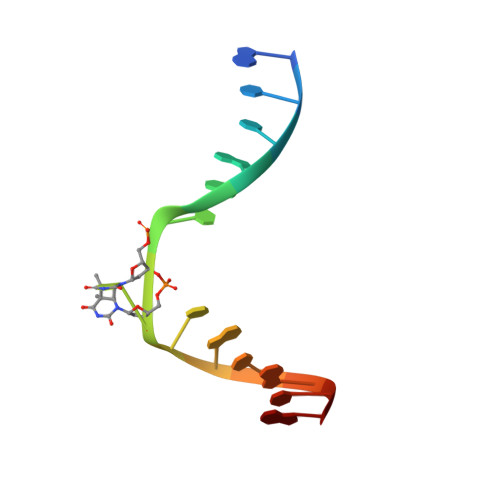
Cyclobutane pyrimidine dimer (CPD) photolyases harness the energy of blue light to repair UV-induced DNA CPDs. Upon binding, CPD photolyases cause the photodamage to flip out of the duplex DNA and into the catalytic site of the enzyme. This process, called base-flipping, induces a kink in the DNA, as well as an unpaired bubble, which are stabilized by a network of protein-nucleic acid interactions. Previously, several co-crystal structures have been reported in which the binding mode of CPD photolyases has been studied in detail. However, in all cases the internucleoside linkage of the photodamage site was a chemically synthesized formacetal analogue and not the natural phosphodiester. Here, the first crystal structure and conformational analysis via molecular-dynamics simulations of a class II CPD photolyase in complex with photodamaged DNA that contains a natural cyclobutane pyrimidine dimer with an intra-lesion phosphodiester linkage are presented. It is concluded that a highly conserved bubble-intruding region (BIR) mediates stabilization of the open form of CPD DNA when complexed with class II CPD photolyases.
Organizational Affiliation:
Institute of Biological Chemistry, Academia Sinica, 128, Academia Road, Sec. 2, Nankang, Taipei 115, Taiwan.