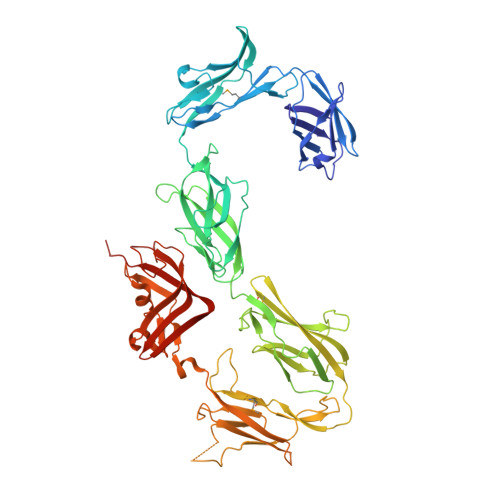
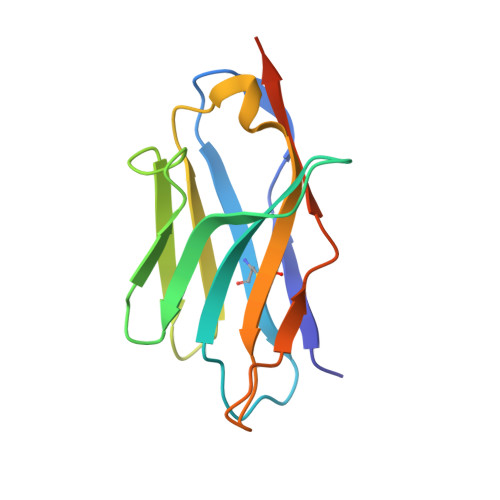
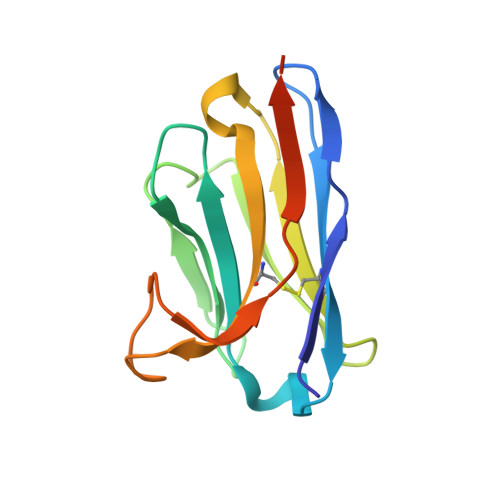
Structure of S-layer protein Sap reveals a mechanism for therapeutic intervention in anthrax.
Fioravanti, A., Van Hauwermeiren, F., Van der Verren, S.E., Jonckheere, W., Goncalves, A., Pardon, E., Steyaert, J., De Greve, H., Lamkanfi, M., Remaut, H.(2019) Nat Microbiol 4: 1805-1814
- PubMed: 31308522
- DOI: https://doi.org/10.1038/s41564-019-0499-1
- Primary Citation of Related Structures:
6HHU, 6QX4 - PubMed Abstract:
Anthrax is an ancient and deadly disease caused by the spore-forming bacterial pathogen Bacillus anthracis. At present, anthrax mostly affects wildlife and livestock, although it remains a concern for human public health-primarily for people who handle contaminated animal products and as a bioterrorism threat due to the high resilience of spores, a high fatality rate of cases and the lack of a civilian vaccination programme 1,2 . The cell surface of B. anthracis is covered by a protective paracrystalline monolayer-known as surface layer or S-layer-that is composed of the S-layer proteins Sap or EA1. Here, we generate nanobodies to inhibit the self-assembly of Sap, determine the structure of the Sap S-layer assembly domain (Sap AD ) and show that the disintegration of the S-layer attenuates the growth of B. anthracis and the pathology of anthrax in vivo. Sap AD comprises six β-sandwich domains that fold and support the formation of S-layers independently of calcium. Sap-inhibitory nanobodies prevented the assembly of Sap and depolymerized existing Sap S-layers in vitro. In vivo, nanobody-mediated disruption of the Sap S-layer resulted in severe morphological defects and attenuated bacterial growth. Subcutaneous delivery of Sap inhibitory nanobodies cleared B. anthracis infection and prevented lethality in a mouse model of anthrax disease. These findings highlight disruption of S-layer integrity as a mechanism that has therapeutic potential in S-layer-carrying pathogens.
- Structural and Molecular Microbiology, Structural Biology Research Center, VIB, Brussels, Belgium. antonella.fioravanti@vub.be.
Organizational Affiliation: