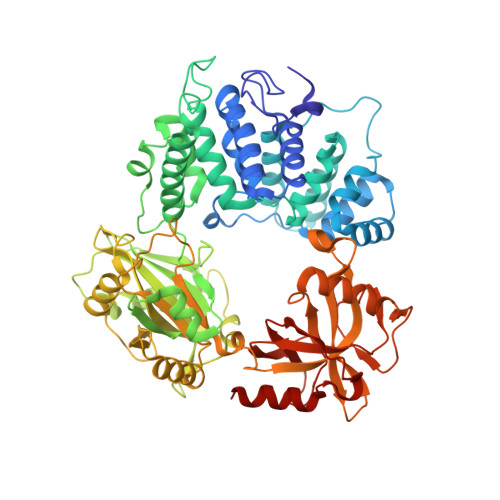
Two-tiered enforcement of high-fidelity DNA ligation.
Tumbale, P.P., Jurkiw, T.J., Schellenberg, M.J., Riccio, A.A., O'Brien, P.J., Williams, R.S.(2019) Nat Commun 10: 5431-5431
- PubMed: 31780661
- DOI: https://doi.org/10.1038/s41467-019-13478-7
- Primary Citation of Related Structures:
6P09, 6P0A, 6P0B, 6P0C, 6P0D, 6P0E, 6Q1V - PubMed Abstract:
DNA ligases catalyze the joining of DNA strands to complete DNA replication, recombination and repair transactions. To protect the integrity of the genome, DNA ligase 1 (LIG1) discriminates against DNA junctions harboring mutagenic 3'-DNA mismatches or oxidative DNA damage, but how such high-fidelity ligation is enforced is unknown. Here, X-ray structures and kinetic analyses of LIG1 complexes with undamaged and oxidatively damaged DNA unveil that LIG1 employs Mg 2+ -reinforced DNA binding to validate DNA base pairing during the adenylyl transfer and nick-sealing ligation reaction steps. Our results support a model whereby LIG1 fidelity is governed by a high-fidelity (HiFi) interface between LIG1, Mg 2+ , and the DNA substrate that tunes the enzyme to release pro-mutagenic DNA nicks. In a second tier of protection, LIG1 activity is surveilled by Aprataxin (APTX), which suppresses mutagenic and abortive ligation at sites of oxidative DNA damage.
- Genome Integrity and Structural Biology Laboratory, National Institute of Environmental Health Sciences, US National Institutes of Health, Department of Health and Human Services, 111 TW Alexander Drive, Research Triangle Park, NC, 27709, USA.
Organizational Affiliation: