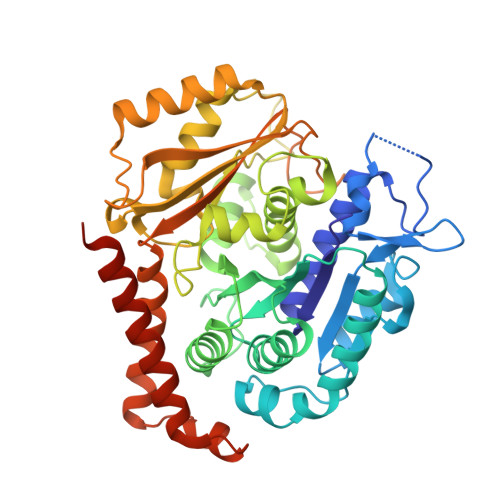
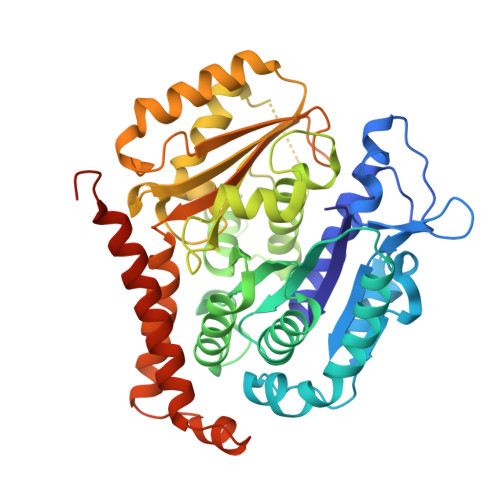
GTP-dependent formation of straight tubulin oligomers leads to microtubule nucleation.
Ayukawa, R., Iwata, S., Imai, H., Kamimura, S., Hayashi, M., Ngo, K.X., Minoura, I., Uchimura, S., Makino, T., Shirouzu, M., Shigematsu, H., Sekimoto, K., Gigant, B., Muto, E.(2021) J Cell Biol 220
- PubMed: 33544140
- DOI: https://doi.org/10.1083/jcb.202007033
- Primary Citation of Related Structures:
6TIS, 6TIU, 6TIY, 6TIZ - PubMed Abstract:
Nucleation of microtubules (MTs) is essential for cellular activities, but its mechanism is unknown because of the difficulty involved in capturing rare stochastic events in the early stage of polymerization. Here, combining rapid flush negative stain electron microscopy (EM) and kinetic analysis, we demonstrate that the formation of straight oligomers of critical size is essential for nucleation. Both GDP and GTP tubulin form single-stranded oligomers with a broad range of curvatures, but upon nucleation, the curvature distribution of GTP oligomers is shifted to produce a minor population of straight oligomers. With tubulin having the Y222F mutation in the β subunit, the proportion of straight oligomers increases and nucleation accelerates. Our results support a model in which GTP binding generates a minor population of straight oligomers compatible with lateral association and further growth to MTs. This study suggests that cellular factors involved in nucleation promote it via stabilization of straight oligomers.
- Laboratory for Molecular Biophysics, RIKEN Center for Brain Science, Saitama, Japan.
Organizational Affiliation: