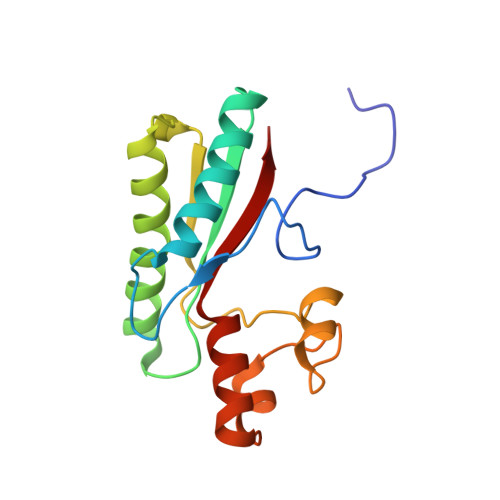
Crystal structure of Type IX secretion system PorE C-terminal domain from Porphyromonas gingivalis in complex with a peptidoglycan fragment.
Trinh, N.T.T., Tran, H.Q., Van Dong, Q., Cambillau, C., Roussel, A., Leone, P.(2020) Sci Rep 10: 7384-7384
- PubMed: 32355178
- DOI: https://doi.org/10.1038/s41598-020-64115-z
- Primary Citation of Related Structures:
6TOP - PubMed Abstract:
Porphyromonas gingivalis, the major human pathogen associated to periodontal diseases, utilizes the Bacteroidetes-specific type IX secretion system (T9SS) to export virulence factors. PorE is a periplasmic multi-domain lipoprotein associated to the outer membrane that was recently identified as essential for T9SS function. Little is known on T9SS at the structural level, and in particular its interaction with peptidoglycan. This prompted us to carry out structural studies on PorE full length as well as on its four isolated domains. Here we report the crystal structure of the C-terminal OmpA_C-like putative peptidoglycan-binding domain at 1.55 Å resolution. An electron density volume was identified in the protein cleft, making it possible to build a naturally-occurring peptidoglycan fragment. This result suggests that PorE interacts with peptidoglycan and that PorE could anchor T9SS to the cell wall.
- Architecture et Fonction des Macromolécules Biologiques, Aix-Marseille Université, UMR 7257, 163 Avenue de Luminy, Case 932, 13009, Marseille, France.
Organizational Affiliation: