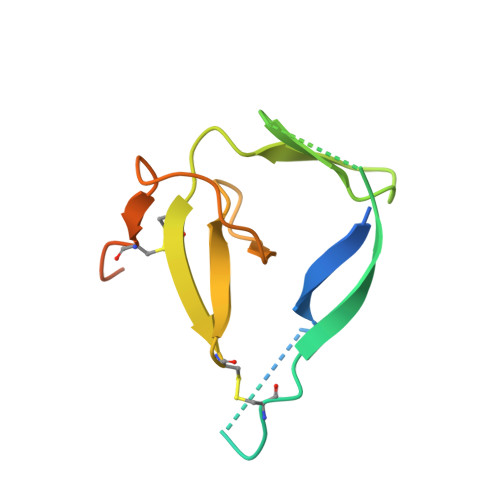
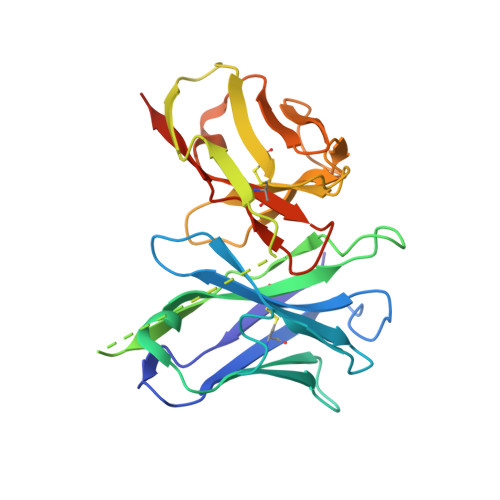
Structure of the prefusion-locking broadly neutralizing antibody RVC20 bound to the rabies virus glycoprotein.
Hellert, J., Buchrieser, J., Larrous, F., Minola, A., de Melo, G.D., Soriaga, L., England, P., Haouz, A., Telenti, A., Schwartz, O., Corti, D., Bourhy, H., Rey, F.A.(2020) Nat Commun 11: 596-596
- PubMed: 32001700
- DOI: https://doi.org/10.1038/s41467-020-14398-7
- Primary Citation of Related Structures:
6TOU - PubMed Abstract:
Rabies virus (RABV) causes fatal encephalitis in more than 59,000 people yearly. Upon the bite of an infected animal, the development of clinical disease can be prevented with post-exposure prophylaxis (PEP), which includes the administration of Rabies immunoglobulin (RIG). However, the high cost and limited availability of serum-derived RIG severely hamper its wide use in resource-limited countries. A safe low-cost alternative is provided by using broadly neutralizing monoclonal antibodies (bnAbs). Here we report the X-ray structure of one of the most potent and most broadly reactive human bnAbs, RVC20, in complex with its target domain III of the RABV glycoprotein (G). The structure reveals that the RVC20 binding determinants reside in a highly conserved surface of G, rationalizing its broad reactivity. We further show that RVC20 blocks the acid-induced conformational change required for membrane fusion. Our results may guide the future development of direct antiviral small molecules for Rabies treatment.
- Structural Virology Unit, Institut Pasteur, CNRS UMR 3569, 25-28 rue du Docteur Roux, Cedex 15, 75724, Paris, France.
Organizational Affiliation: