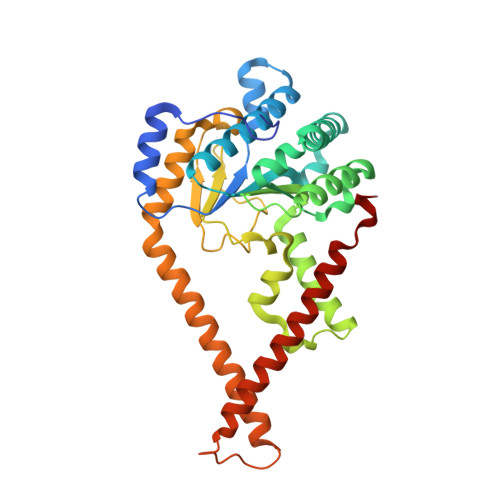
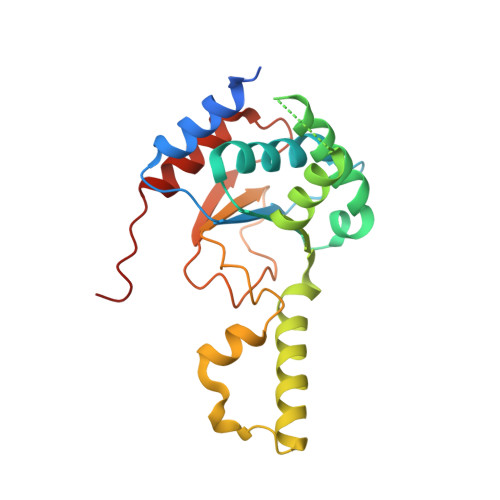
Structural basis for long-chain isoprenoid synthesis by cis -prenyltransferases.
Giladi, M., Lisnyansky Bar-El, M., Vankova, P., Ferofontov, A., Melvin, E., Alkaderi, S., Kavan, D., Redko, B., Haimov, E., Wiener, R., Man, P., Haitin, Y.(2022) Sci Adv 8: eabn1171-eabn1171
- PubMed: 35584224
- DOI: https://doi.org/10.1126/sciadv.abn1171
- Primary Citation of Related Structures:
7PAX, 7PAY, 7PB0, 7PB1 - PubMed Abstract:
Isoprenoids are synthesized by the prenyltransferase superfamily, which is subdivided according to the product stereoisomerism and length. In short- and medium-chain isoprenoids, product length correlates with active site volume. However, enzymes synthesizing long-chain products and rubber synthases fail to conform to this paradigm, because of an unexpectedly small active site. Here, we focused on the human cis -prenyltransferase complex (h cis -PT), residing at the endoplasmic reticulum membrane and playing a crucial role in protein glycosylation. Crystallographic investigation of h cis -PT along the reaction cycle revealed an outlet for the elongating product. Hydrogen-deuterium exchange mass spectrometry analysis showed that the hydrophobic active site core is flanked by dynamic regions consistent with separate inlet and outlet orifices. Last, using a fluorescence substrate analog, we show that product elongation and membrane association are closely correlated. Together, our results support direct membrane insertion of the elongating isoprenoid during catalysis, uncoupling active site volume from product length.
- Department of Physiology and Pharmacology, Sackler School of Medicine, Tel-Aviv University, Tel-Aviv 6997801, Israel.
Organizational Affiliation: