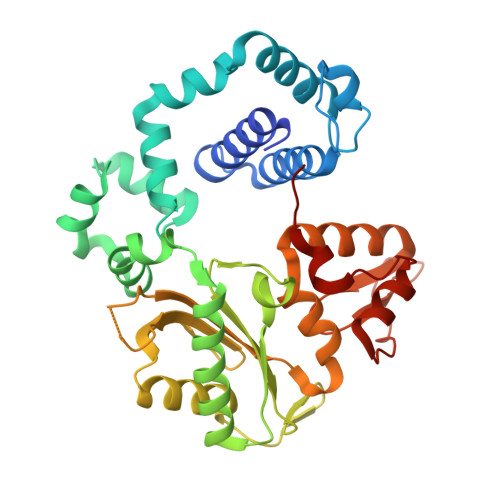
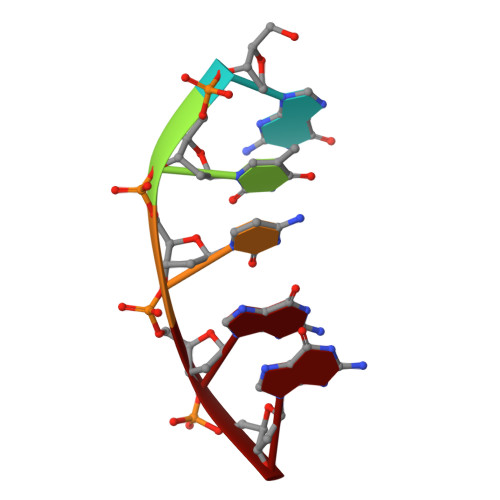
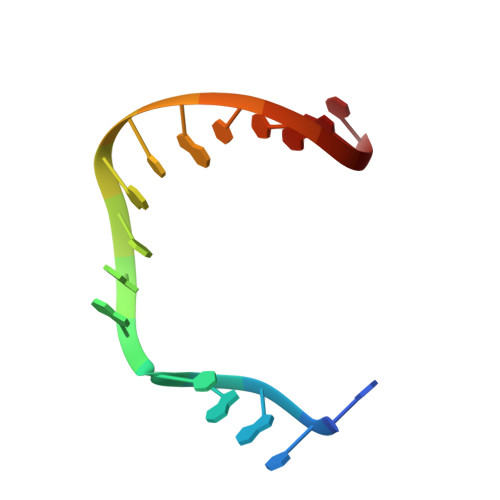
Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Kumar, A., Reed, A.J., Zahurancik, W.J., Daskalova, S.M., Hecht, S.M., Suo, Z.(2022) Proc Natl Acad Sci U S A 119: e2118940119-e2118940119
- PubMed: 35238634
- DOI: https://doi.org/10.1073/pnas.2118940119
- Primary Citation of Related Structures:
7RBE, 7RBF, 7RBG, 7RBH, 7RBI, 7RBJ, 7RBK, 7RBL, 7RBM, 7RBN, 7RBO - PubMed Abstract:
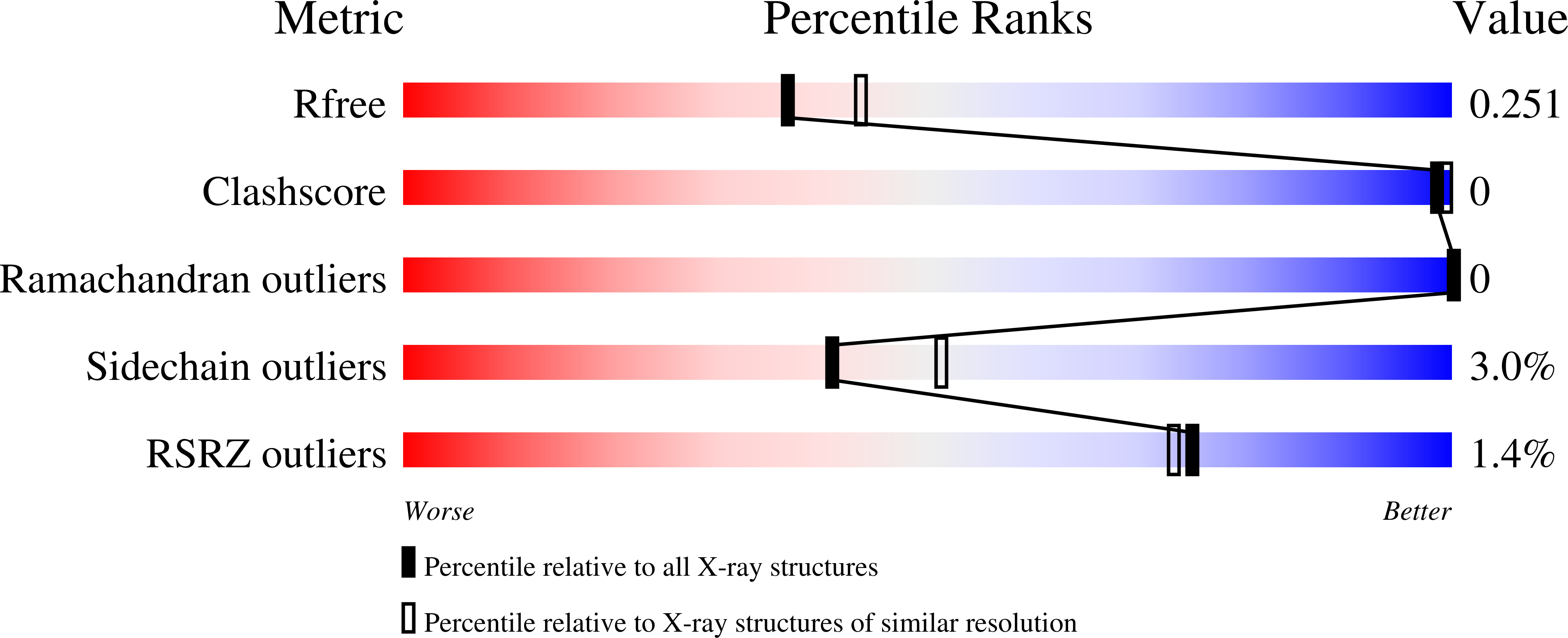
SignificanceBase excision repair (BER) is one of the major DNA repair pathways used to fix a myriad of cellular DNA lesions. The enzymes involved in BER, including DNA polymerase β (Polβ), have been identified and characterized, but how they act together to efficiently perform BER has not been fully understood. Through gel electrophoresis, mass spectrometry, and kinetic analysis, we discovered that the two enzymatic activities of Polβ can be interlocked, rather than functioning independently from each other, when processing DNA intermediates formed in BER. The finding prompted us to hypothesize a modified BER pathway. Through conventional and time-resolved X-ray crystallography, we solved 11 high-resolution crystal structures of cross-linked Polβ complexes and proposed a detailed chemical mechanism for Polβ's 5'-deoxyribose-5-phosphate lyase activity.
Organizational Affiliation:
Department of Biomedical Sciences, Florida State University College of Medicine, Tallahassee, FL 32306.