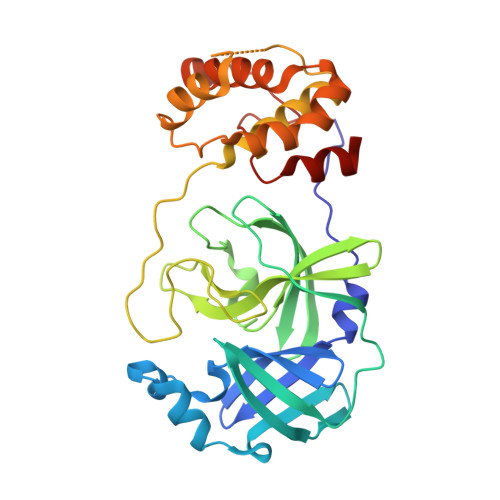
An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Sun, Z., Wang, L., Li, X., Fan, C., Xu, J., Shi, Z., Qiao, H., Lan, Z., Zhang, X., Li, L., Zhou, X., Geng, Y.(2022) Proc Natl Acad Sci U S A 119
- PubMed: 35324337
- DOI: https://doi.org/10.1073/pnas.2120913119
- Primary Citation of Related Structures:
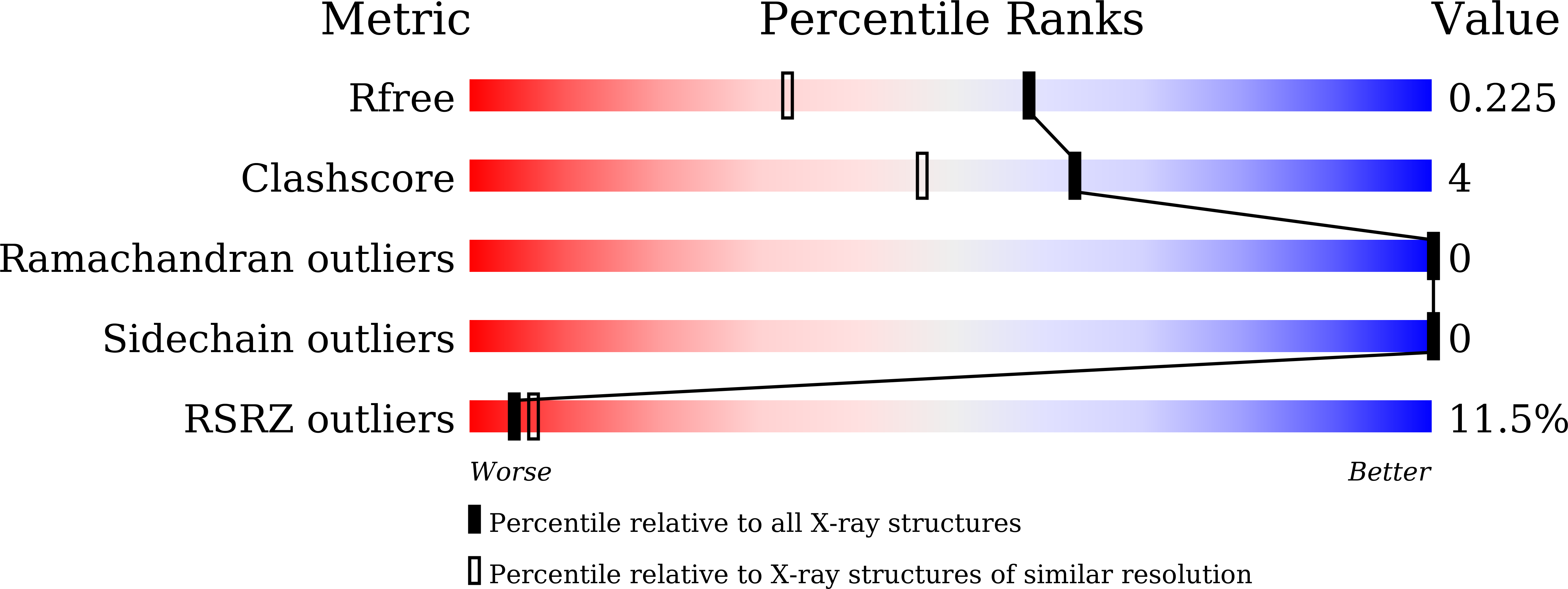
7VFA, 7VFB - PubMed Abstract:
SignificanceThe coronavirus main protease (M pro ) is required for viral replication. Here, we obtained the extended conformation of the native monomer of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) M pro by trapping it with nanobodies and found that the catalytic domain and the helix domain dissociate, revealing allosteric targets. Another monomeric state is termed compact conformation and is similar to one protomer of the dimeric form. We designed a Nanoluc Binary Techonology (NanoBiT)-based high-throughput allosteric inhibitor assay based on structural conformational change. Our results provide insight into the maturation, dimerization, and catalysis of the coronavirus M pro and pave a way to develop an anticoronaviral drug through targeting the maturation process to inhibit the autocleavage of M pro .
Organizational Affiliation:
The Chinese Academy of Sciences Key Laboratory of Receptor Research, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China.