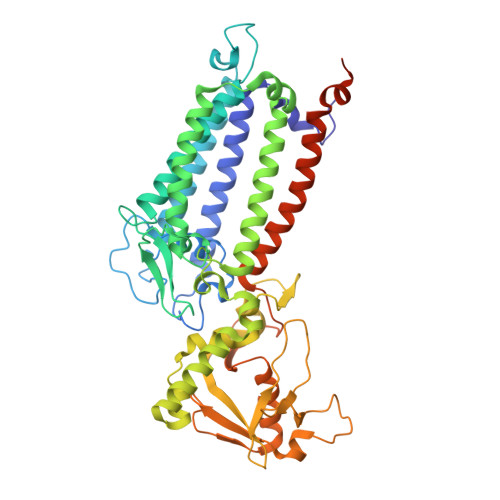
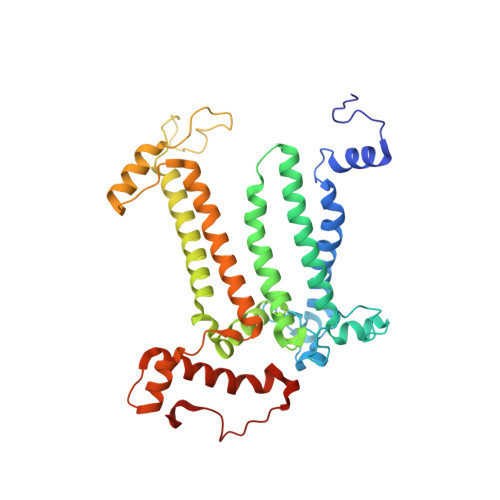
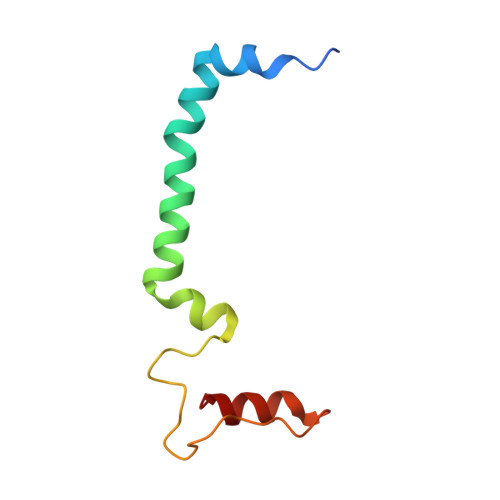
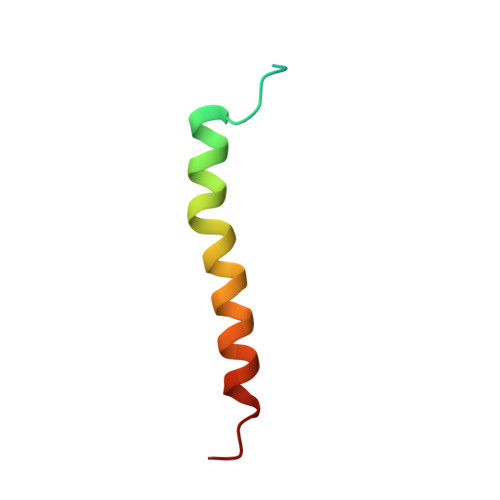
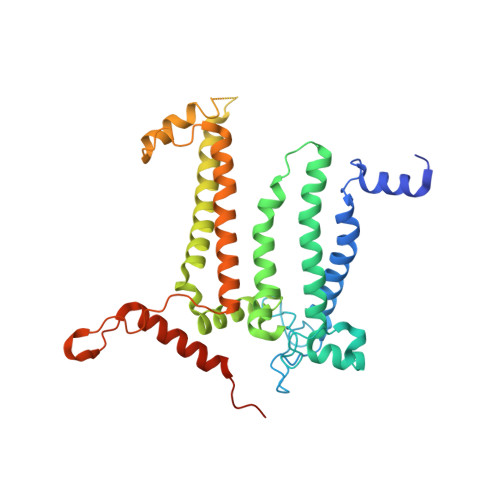
Structural basis for an early stage of the photosystem II repair cycle in Chlamydomonas reinhardtii.
Li, A., You, T., Pang, X., Wang, Y., Tian, L., Li, X., Liu, Z.(2024) Nat Commun 15: 5211-5211
- PubMed: 38890314
- DOI: https://doi.org/10.1038/s41467-024-49532-2
- Primary Citation of Related Structures:
8KDE, 8ZEE - PubMed Abstract:
Photosystem II (PSII) catalyzes water oxidation and plastoquinone reduction by utilizing light energy. It is highly susceptible to photodamage under high-light conditions and the damaged PSII needs to be restored through a process known as the PSII repair cycle. The detailed molecular mechanism underlying the PSII repair process remains mostly elusive. Here, we report biochemical and structural features of a PSII-repair intermediate complex, likely arrested at an early stage of the PSII repair process in the green alga Chlamydomonas reinhardtii. The complex contains three protein factors associated with a damaged PSII core, namely Thylakoid Enriched Factor 14 (TEF14), Photosystem II Repair Factor 1 (PRF1), and Photosystem II Repair Factor 2 (PRF2). TEF14, PRF1 and PRF2 may facilitate the release of the manganese-stabilizing protein PsbO, disassembly of peripheral light-harvesting complexes from PSII and blockage of the Q B site, respectively. Moreover, an α-tocopherol quinone molecule is located adjacent to the heme group of cytochrome b 559 , potentially fulfilling a photoprotective role by preventing the generation of reactive oxygen species.
- Key Laboratory of Biomacromolecules (CAS), National Laboratory of Biomacromolecules, CAS Centre for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, 100101, China.
Organizational Affiliation: