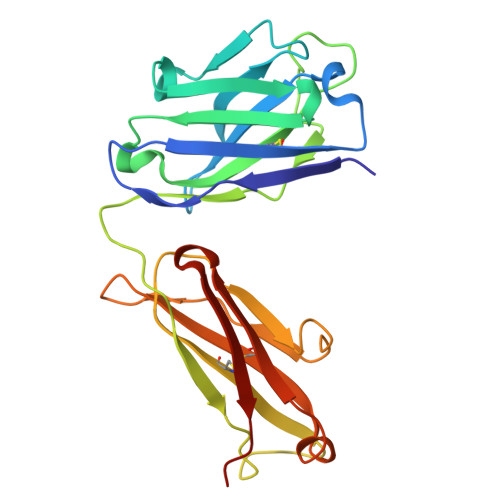
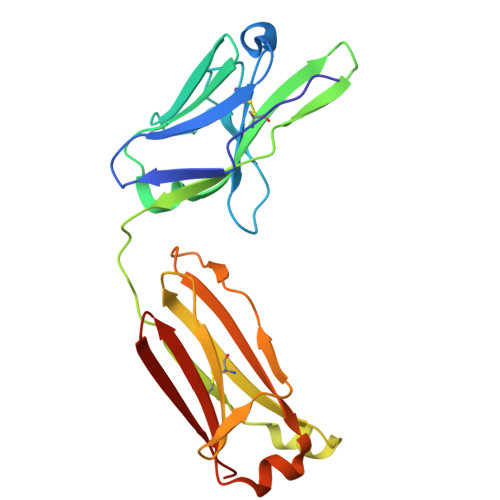
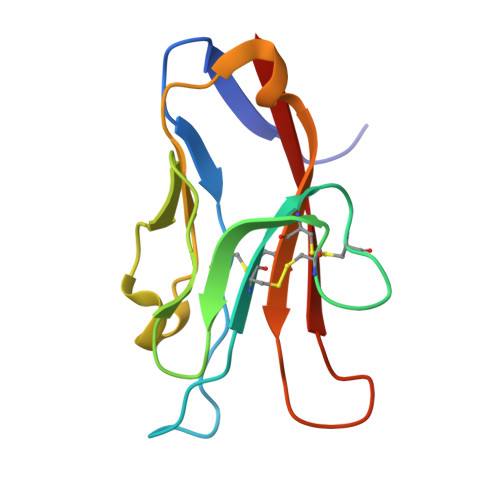
Generation of AZD7789, a novel PD-1 and TIM-3 targeting bispecific antibody, which binds to a differentiated epitope of TIM-3
Clancy-Thompson, E., Perry, T., Pryts, S., Jaiswal, A., Oganesyan, V., Moynihan, J., van Dyk, N., Yang, C., Garcia, A., Yu, J., Miller, C., Mulgrew, K., Cobbold, M., Mazor, Y., Hammond, S.A., Pollizzi, K.N.To be published.