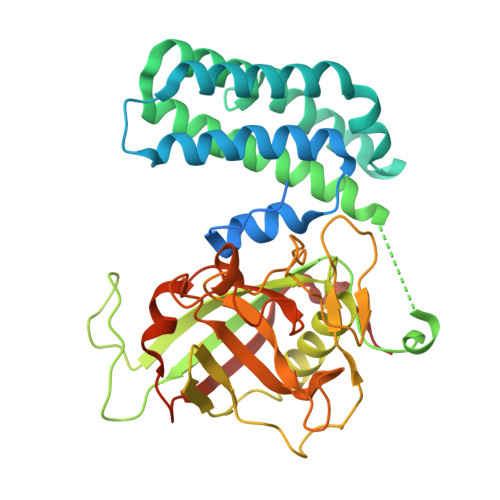
Crystal structure of cyclophilin 37 from Arabidopsis thaliana.
Han, X., Jiang, J., Lu, Z., Bai, J., Qin, X., Dong, S.(2025) Acta Crystallogr F Struct Biol Commun 81: 171-176
- PubMed: 40091855
- DOI: https://doi.org/10.1107/S2053230X25001979
- Primary Citation of Related Structures:
9LH4 - PubMed Abstract:
Photosynthesis is the largest-scale energy and material conversion process on Earth. The cytchrome (Cyt) b 6 f complex plays a crucial role in photosynthesis. Under high-light conditions, cyclophilin 37 (CYP37) in Arabidopsis thaliana (AtCYP37) can interact with the PetA subunit of Cyt b 6 f, thereby helping plants initiate photoprotection. Here, we purified, crystallized and determined a 1.95 Å resolution structure of AtCYP37. Overall, AtCYP37 consists of an N-terminal domain dominated by α-helices and a C-terminal domain mainly composed of β-strands and random coils. The structure shows significant similarity to those of Anabaena sp. CYPA and A. thaliana CYP38. Understanding the structure of AtCYP37 is significant as it may help to decipher how plants regulate photosynthesis and protect against high light damage, contributing to a broader understanding of plant photobiology and potentially guiding future research in improving plant stress tolerance.
- School of Biological Science and Technology, University of Jinan, Jinan, Shandong 250022, People's Republic of China.
Organizational Affiliation: