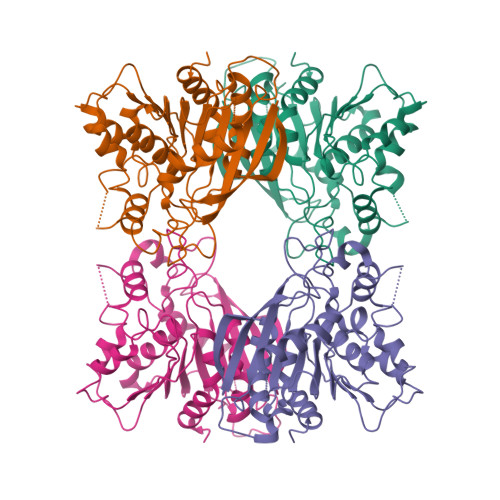
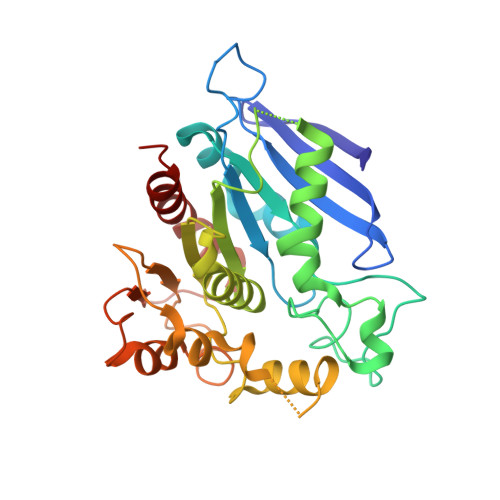
Structural characterization and reversal of the natural organophosphate resistance of a D-type esterase, Saccharomyces cerevisiae S-formylglutathione hydrolase.
Legler, P.M., Kumaran, D., Swaminathan, S., Studier, F.W., Millard, C.B.(2008) Biochemistry 47: 9592-9601
- PubMed: 18707125
- DOI: https://doi.org/10.1021/bi8010016
- Primary Citation of Related Structures:
1PV1, 3C6B - PubMed Abstract:
Saccharomyces cerevisiae expresses a 67.8 kDa homodimeric serine thioesterase, S-formylglutathione hydrolase (SFGH), that is 39.9% identical with human esterase D. Both enzymes possess significant carboxylesterase and S-formylglutathione thioesterase activity but are unusually resistant to organophosphate (OP) inhibitors. We determined the X-ray crystal structure of yeast (y) SFGH to 2.3 A resolution by multiwavelength anomalous dispersion and used the structure to guide site-specific mutagenesis experiments addressing substrate and inhibitor reactivity. Our results demonstrate a steric mechanism of OP resistance mediated by a single indole ring (W197) located in an enzyme "acyl pocket". The W197I substitution enhances ySFGH reactivity with paraoxon by >1000-fold ( k i (W197I) = 16 +/- 2 mM (-1) h (-1)), thereby overcoming natural OP resistance. W197I increases the rate of OP inhibition under pseudo-first-order conditions but does not accelerate OP hydrolysis. The structure of the paraoxon-inhibited W197I variant was determined by molecular replacement (2.2 A); it revealed a stabilized sulfenic acid at Cys60. Wild-type (WT) ySFGH is inhibited by thiol reactive compounds and is sensitive to oxidation; thus, the cysteine sulfenic acid may play a role in the regulation of a "D-type" esterase. The structure of the W197I variant is the first reported cysteine sulfenic acid in a serine esterase. We constructed five Cys60/W197I variants and show that introducing a positive charge near the oxyanion hole, W197I/C60R or W197I/C60K, results in a further enhancement of the rates of phosphorylation with paraoxon ( k i = 42 or 80 mM (-1) h (-1), respectively) but does not affect the dephosphorylation of the enzyme. We also characterized three histidine substitutions near the oxyanion hole, G57H, L58H, and M162H, which significantly decrease esterase activity.
Organizational Affiliation:
Division of Biochemistry, Walter Reed Army Institute of Research, Silver Spring, Maryland 20910, USA.