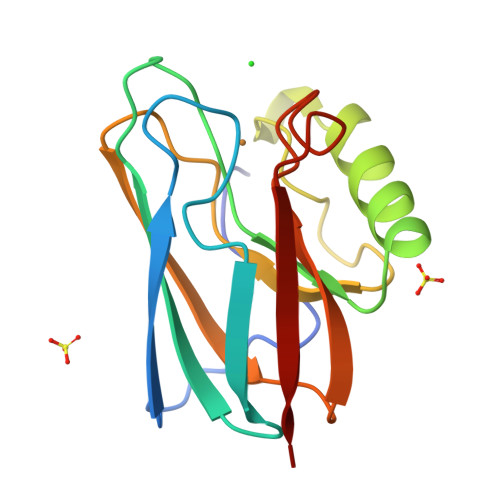
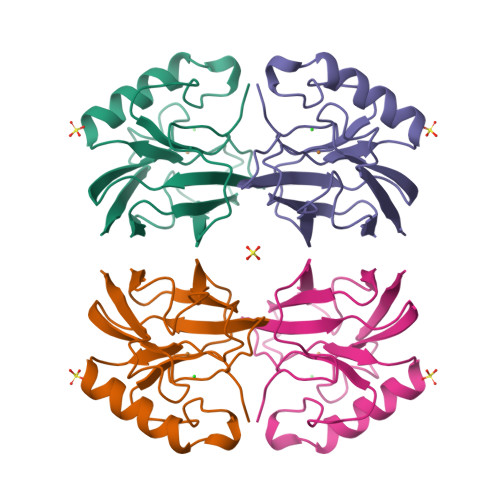
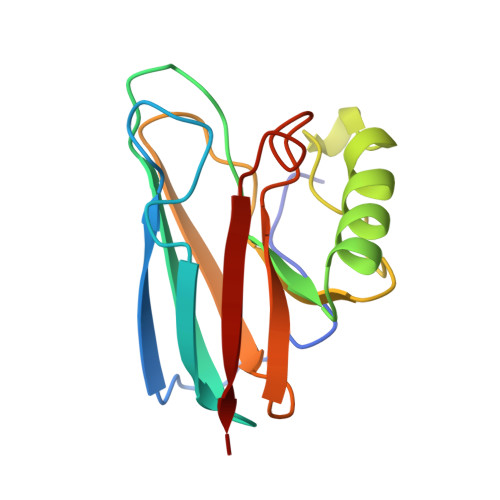
Crystal structure of auracyanin, a "blue" copper protein from the green thermophilic photosynthetic bacterium Chloroflexus aurantiacus.
Bond, C.S., Blankenship, R.E., Freeman, H.C., Guss, J.M., Maher, M.J., Selvaraj, F.M., Wilce, M.C., Willingham, K.M.(2001) J Mol Biology 306: 47-67
- PubMed: 11178893
- DOI: https://doi.org/10.1006/jmbi.2000.4201
- Primary Citation of Related Structures:
1QHQ - PubMed Abstract:
Auracyanin B, one of two similar blue copper proteins produced by the thermophilic green non-sulfur photosynthetic bacterium Chloroflexus aurantiacus, crystallizes in space group P6(4)22 (a=b=115.7 A, c=54.6 A). The structure was solved using multiple wavelength anomalous dispersion data recorded about the CuK absorption edge, and was refined at 1.55 A resolution. The molecular model comprises 139 amino acid residues, one Cu, 247 H(2)O molecules, one Cl(-) and two SO(4)(2-). The final residual and estimated standard uncertainties are R=0.198, ESU=0.076 A for atomic coordinates and ESU=0.05 A for Cu---ligand bond lengths, respectively. The auracyanin B molecule has a standard cupredoxin fold. With the exception of an additional N-terminal strand, the molecule is very similar to that of the bacterial cupredoxin, azurin. As in other cupredoxins, one of the Cu ligands lies on strand 4 of the polypeptide, and the other three lie along a large loop between strands 7 and 8. The Cu site geometry is discussed with reference to the amino acid spacing between the latter three ligands. The crystallographically characterized Cu-binding domain of auracyanin B is probably tethered to the periplasmic side of the cytoplasmic membrane by an N-terminal tail that exhibits significant sequence identity with known tethers in several other membrane-associated electron-transfer proteins.
Organizational Affiliation:
Department of Biochemistry, University of Sydney, New South Wales, 2006, Australia.