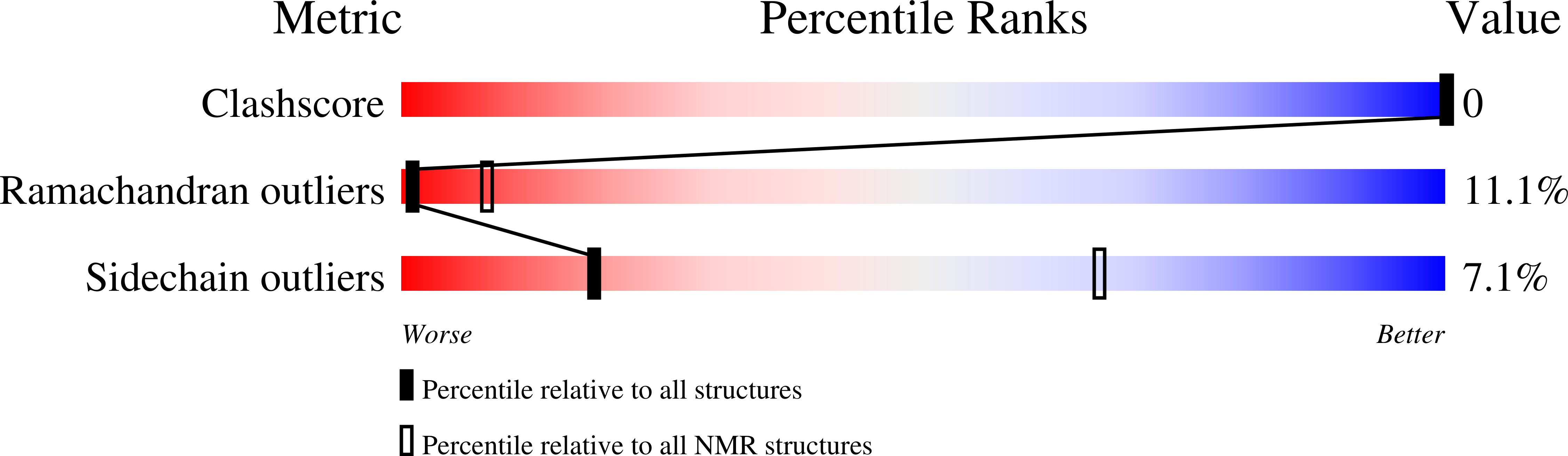Supramolecular structure of membrane-associated polypeptides by combining solid-state NMR and molecular dynamics simulations.
Weingarth, M., Ader, C., Melquiond, A.J., Nand, D., Pongs, O., Becker, S., Bonvin, A.M., Baldus, M.(2012) Biophys J 103: 29-37
- PubMed: 22828329
- DOI: https://doi.org/10.1016/j.bpj.2012.05.016
- Primary Citation of Related Structures:
2LNY - PubMed Abstract:
Elemental biological functions such as molecular signal transduction are determined by the dynamic interplay between polypeptides and the membrane environment. Determining such supramolecular arrangements poses a significant challenge for classical structural biology methods. We introduce an iterative approach that combines magic-angle spinning solid-state NMR spectroscopy and atomistic molecular dynamics simulations for the determination of the structure and topology of membrane-bound systems with a resolution and level of accuracy difficult to obtain by either method alone. Our study focuses on the Shaker B ball peptide that is representative for rapid N-type inactivating domains of voltage-gated K(+) channels, associated with negatively charged lipid bilayers.
Organizational Affiliation:
Bijvoet Center for Biomolecular Research, Utrecht University, Utrecht, The Netherlands.