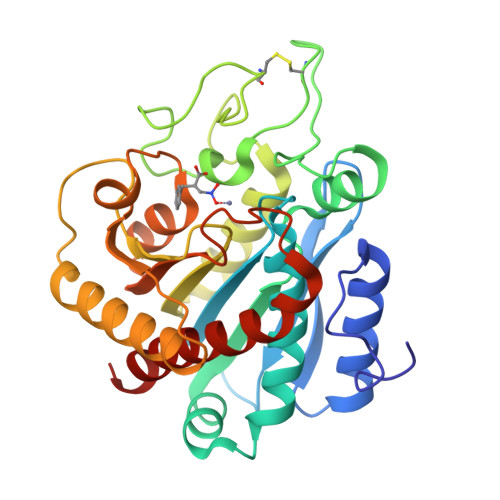
Nitro as a novel zinc-binding group in the inhibition of carboxypeptidase A
Wang, S.-H., Wang, S.-F., Xuan, W., Zeng, Z.-H., Jin, J.-Y., Ma, J., Tian, G.R.(2008) Bioorg Med Chem 16: 3596-3601
- PubMed: 18289863
- DOI: https://doi.org/10.1016/j.bmc.2008.02.010
- Primary Citation of Related Structures:
2RFH - PubMed Abstract:
2-Substituted 3-nitropropanoic acids were designed and synthesized as inhibitors against carboxypeptidase A (CPA). (R)-2-Benzyl- 3-nitropropanoic acid showed a potent inhibition against CPA (K(i)=0.15 microM). X-ray crystallography discloses that the nitro group well mimics the transition state occurred in the hydrolysis catalyzed by CPA, that is, an O,O'-bidentate coordination to the zinc ion and the two respective hydrogen bonds with Glu-270 and Arg-127. Because the nitro group is a planar species, we proposed (R)-2-benzyl-3-nitropropanoic acid as a pseudo-transition-state analog inhibitor against CPA.
Organizational Affiliation:
Department of Chemistry, Yanbian University, Yanji, Jilin 133002, China.