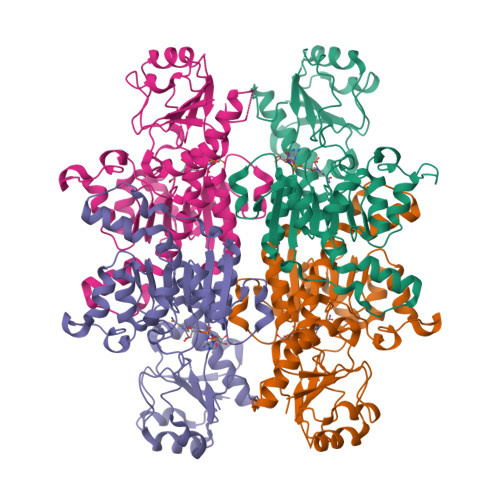
Structural Characterization of D-Isomer Specific 2-Hydroxyacid Dehydrogenase from Lactobacillus Delbrueckii Ssp. Bulgaricus
Holton, S.J., Anandhakrishnan, M., Geerlof, A., Wilmanns, M.(2013) J Struct Biol 181: 179
- PubMed: 23110853
- DOI: https://doi.org/10.1016/j.jsb.2012.10.009
- Primary Citation of Related Structures:
2YQ4, 2YQ5 - PubMed Abstract:
Hydroxyacid dehydrogenases, responsible for the stereospecific conversion of 2-keto acids to 2-hydroxyacids in lactic acid producing bacteria, have a range of biotechnology applications including antibiotic synthesis, flavor development in dairy products and the production of valuable synthons. The genome of Lactobacillus delbrueckii ssp. bulgaricus, a member of the heterogeneous group of lactic acid bacteria, encodes multiple hydroxyacid dehydrogenases whose structural and functional properties remain poorly characterized. Here, we report the apo and coenzyme NAD⁺ complexed crystal structures of the L. bulgaricusD-isomer specific 2-hydroxyacid dehydrogenase, D2-HDH. Comparison with closely related members of the NAD-dependent dehydrogenase family reveals that whilst the D2-HDH core fold is structurally conserved, the substrate-binding site has a number of non-canonical features that may influence substrate selection and thus dictate the physiological function of the enzyme.
Organizational Affiliation:
EMBL c/o DESY, Notkestrasse 85, D-22603 Hamburg, Germany. sjholton@googlemail.com