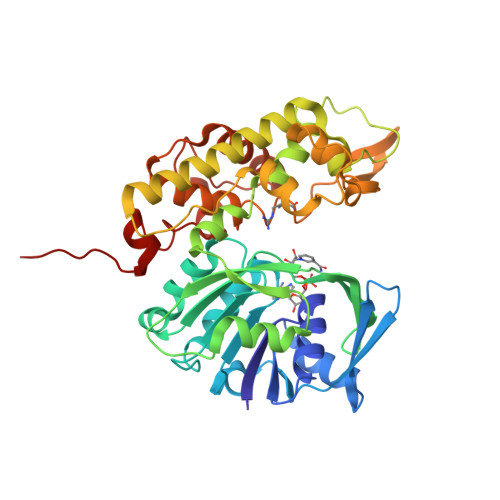
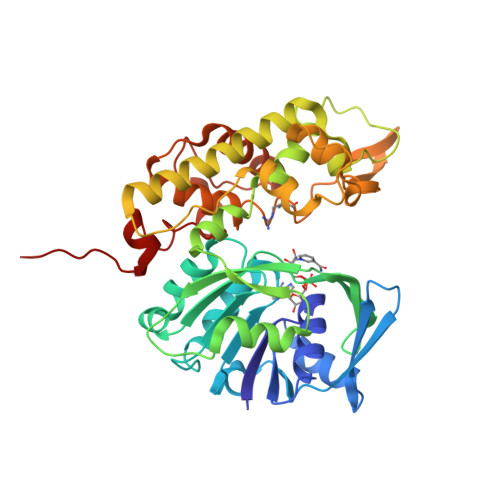
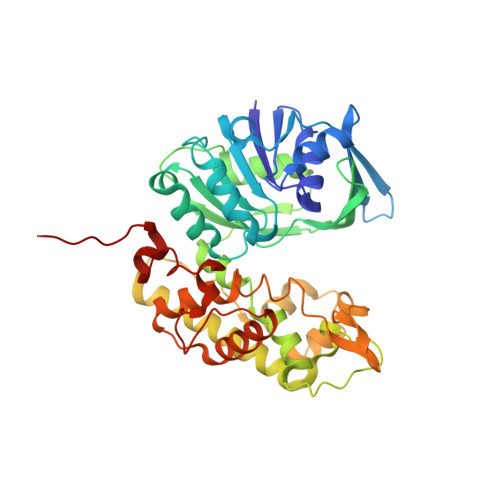
A structural basis for substrate selectivity and stereoselectivity in octopine dehydrogenase from Pecten maximus.
Smits, S.H., Mueller, A., Schmitt, L., Grieshaber, M.K.(2008) J Mol Biology 381: 200-211
- PubMed: 18599075
- DOI: https://doi.org/10.1016/j.jmb.2008.06.003
- Primary Citation of Related Structures:
3C7A, 3C7C, 3C7D - PubMed Abstract:
Octopine dehydrogenase [N(2)-(D-1-carboxyethyl)-L-arginine:NAD(+) oxidoreductase] (OcDH) from the adductor muscle of the great scallop Pecten maximus catalyzes the reductive condensation of l-arginine and pyruvate to octopine during escape swimming. This enzyme, which is a prototype of opine dehydrogenases (OpDHs), oxidizes glycolytically born NADH to NAD(+), thus sustaining anaerobic ATP provision during short periods of strenuous muscular activity. In contrast to some other OpDHs, OcDH uses only l-arginine as the amino acid substrate. Here, we report the crystal structures of OcDH in complex with NADH and the binary complexes NADH/l-arginine and NADH/pyruvate, providing detailed information about the principles of substrate recognition, ligand binding and the reaction mechanism. OcDH binds its substrates through a combination of electrostatic forces and size selection, which guarantees that OcDH catalysis proceeds with substrate selectivity and stereoselectivity, giving rise to a second chiral center and exploiting a "molecular ruler" mechanism.
Organizational Affiliation:
Institute of Biochemistry, Heinrich Heine University, Universitaetsstrasse 1, 40225 Duesseldorf, Germany.