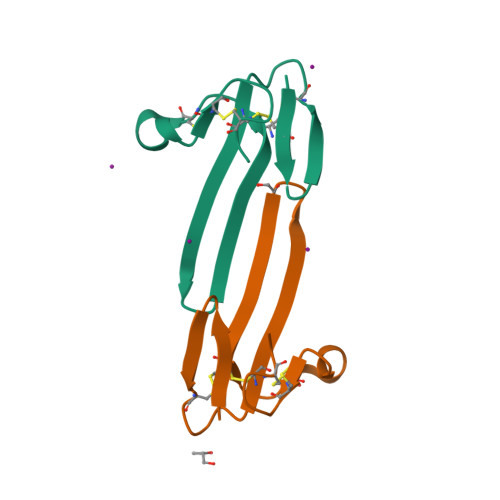
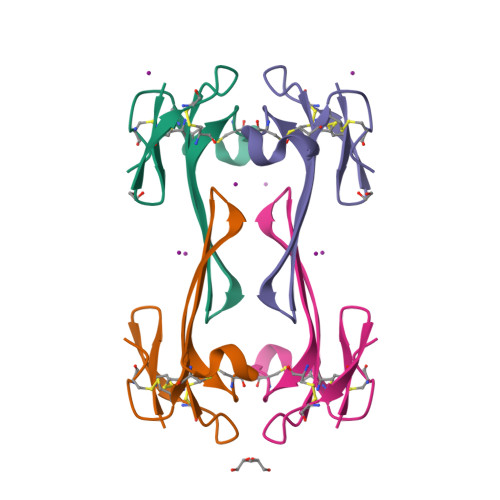
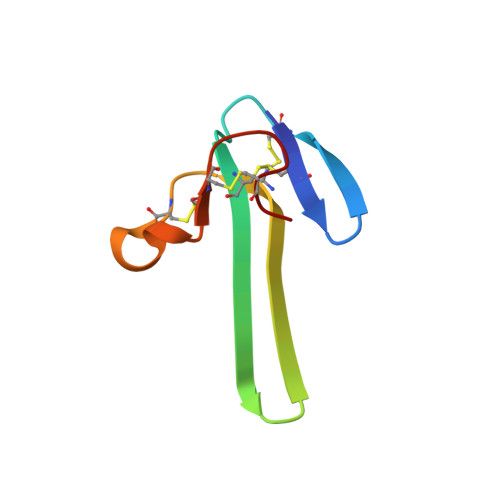
Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition.
Mourier, G., Salinas, M., Kessler, P., Stura, E.A., Leblanc, M., Tepshi, L., Besson, T., Diochot, S., Baron, A., Douguet, D., Lingueglia, E., Servent, D.(2016) J Biological Chem 291: 2616-2629
- PubMed: 26680001
- DOI: https://doi.org/10.1074/jbc.M115.702373
- Primary Citation of Related Structures:
5DO6, 5DU1, 5DZ5 - PubMed Abstract:
Mambalgins are peptides isolated from mamba venom that specifically inhibit a set of acid-sensing ion channels (ASICs) to relieve pain. We show here the first full stepwise solid phase peptide synthesis of mambalgin-1 and confirm the biological activity of the synthetic toxin both in vitro and in vivo. We also report the determination of its three-dimensional crystal structure showing differences with previously described NMR structures. Finally, the functional domain by which the toxin inhibits ASIC1a channels was identified in its loop II and more precisely in the face containing Phe-27, Leu-32, and Leu-34 residues. Moreover, proximity between Leu-32 in mambalgin-1 and Phe-350 in rASIC1a was proposed from double mutant cycle analysis. These data provide information on the structure and on the pharmacophore for ASIC channel inhibition by mambalgins that could have therapeutic value against pain and probably other neurological disorders.
Organizational Affiliation:
From the Commissariat à l'Energie Atomique, iBiTecS, Service d'Ingénierie Moléculaire des Protéines, 91191 Gif-sur-Yvette.