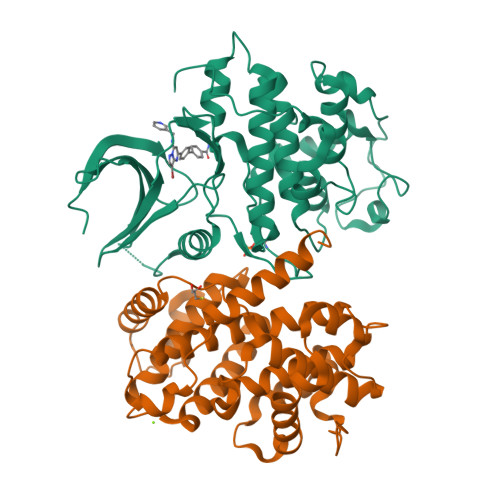
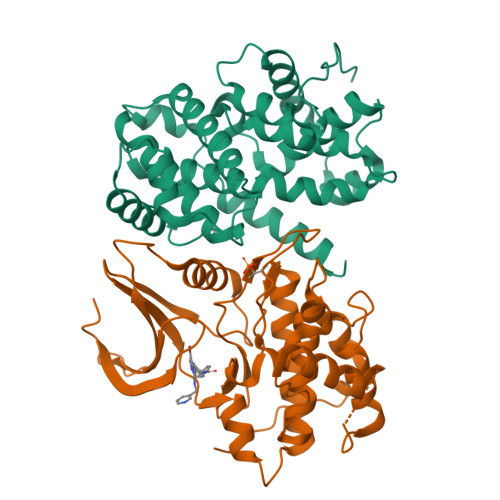
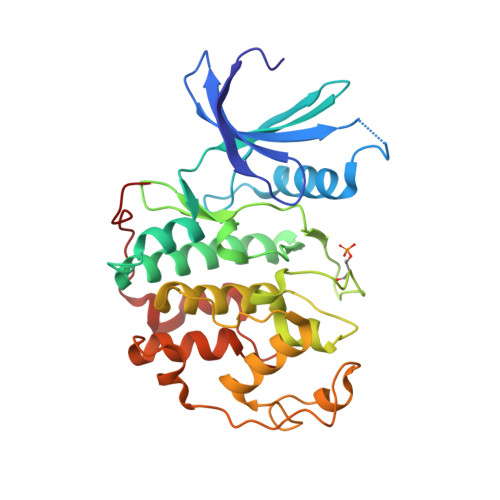
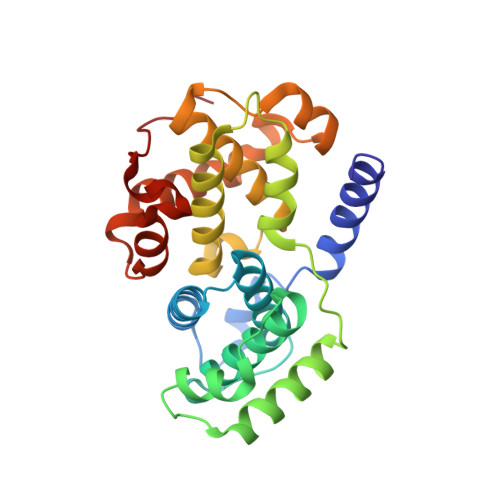
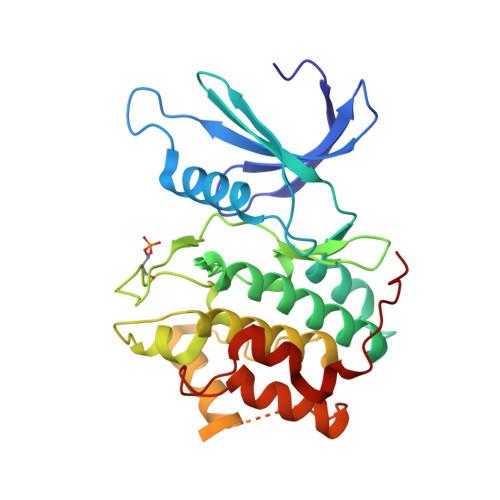
Explicit treatment of active-site waters enhances quantum mechanical/implicit solvent scoring: Inhibition of CDK2 by new pyrazolo[1,5-a]pyrimidines.
Hylsova, M., Carbain, B., Fanfrlik, J., Musilova, L., Haldar, S., Kopruluoglu, C., Ajani, H., Brahmkshatriya, P.S., Jorda, R., Krystof, V., Hobza, P., Echalier, A., Paruch, K., Lepsik, M.(2016) Eur J Med Chem 126: 1118-1128
- PubMed: 28039837
- DOI: https://doi.org/10.1016/j.ejmech.2016.12.023
- Primary Citation of Related Structures:
5LMK - PubMed Abstract:
We present comprehensive testing of solvent representation in quantum mechanics (QM)-based scoring of protein-ligand affinities. To this aim, we prepared 21 new inhibitors of cyclin-dependent kinase 2 (CDK2) with the pyrazolo[1,5-a]pyrimidine core, whose activities spanned three orders of magnitude. The crystal structure of a potent inhibitor bound to the active CDK2/cyclin A complex revealed that the biphenyl substituent at position 5 of the pyrazolo[1,5-a]pyrimidine scaffold was located in a previously unexplored pocket and that six water molecules resided in the active site. Using molecular dynamics, protein-ligand interactions and active-site water H-bond networks as well as thermodynamics were probed. Thereafter, all the inhibitors were scored by the QM approach utilizing the COSMO implicit solvent model. Such a standard treatment failed to produce a correlation with the experiment (R 2 = 0.49). However, the addition of the active-site waters resulted in significant improvement (R 2 = 0.68). The activities of the compounds could thus be interpreted by taking into account their specific noncovalent interactions with CDK2 and the active-site waters. In summary, using a combination of several experimental and theoretical approaches we demonstrate that the inclusion of explicit solvent effects enhance QM/COSMO scoring to produce a reliable structure-activity relationship with physical insights. More generally, this approach is envisioned to contribute to increased accuracy of the computational design of novel inhibitors.
Organizational Affiliation:
Department of Chemistry, CZ Openscreen, Masaryk University, Kamenice 5, 625 00 Brno, Czech Republic.