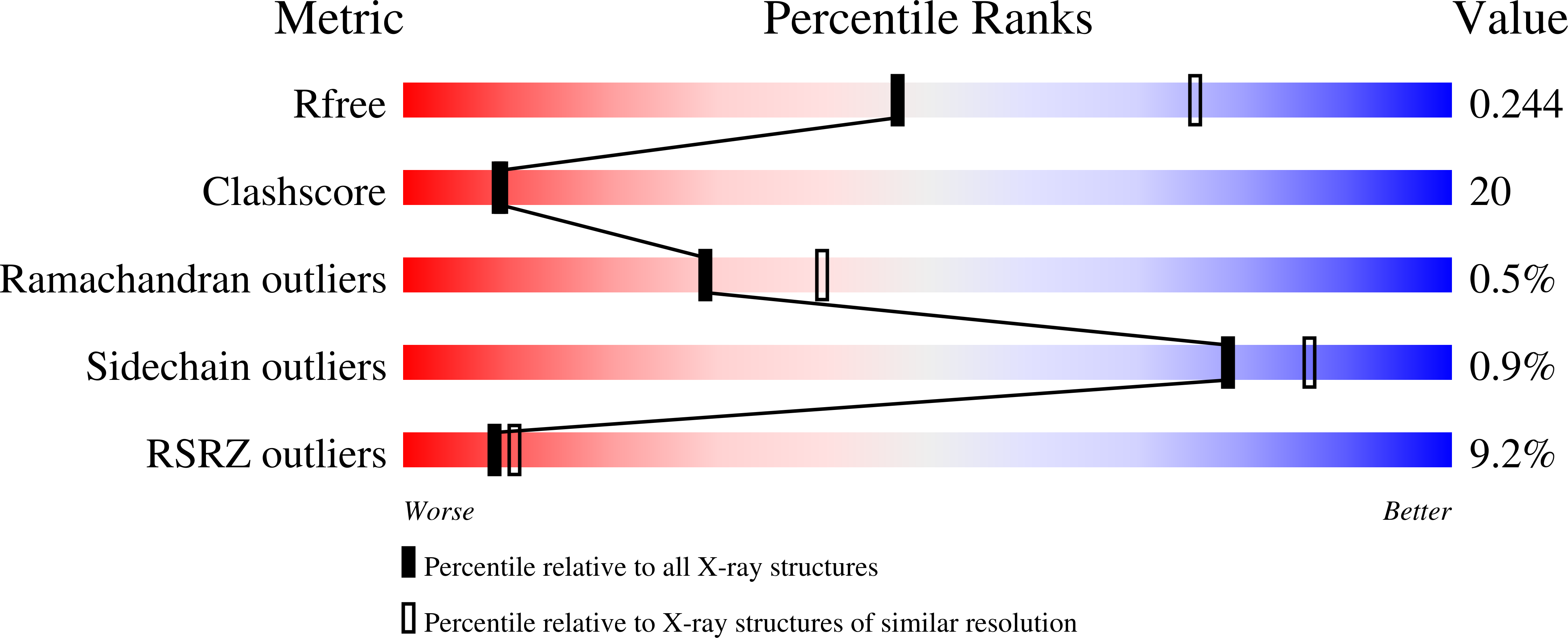
Hit-to-lead optimization and discovery of a potent, and orally bioavailable G protein coupled receptor kinase 2 (GRK2) inhibitor.
Xu, G., Gaul, M.D., Liu, Z., DesJarlais, R.L., Qi, J., Wang, W., Krosky, D., Petrounia, I., Milligan, C.M., Hermans, A., Lu, H.R., Huang, D.Z., Xu, J.Z., Spurlino, J.C.(2020) Bioorg Med Chem Lett 30: 127602-127602
- PubMed: 33038544
- DOI: https://doi.org/10.1016/j.bmcl.2020.127602
- Primary Citation of Related Structures:
7K7L, 7K7Z - PubMed Abstract:
G-protein coupled receptor kinase 2 (GRK2), which is upregulated in the failing heart, appears to play a critical role in heart failure (HF) progression in part because enhanced GRK2 activity promotes dysfunction of β-adrenergic signaling and myocyte death. An orally bioavailable GRK2 inhibitor could offer unique therapeutic outcomes that cannot be attained by current heart failure treatments that directly target GPCRs or angiotensin-converting enzyme. Herein, we describe the discovery of a potent, selective, and orally bioavailable GRK2 inhibitor, 8h, through high-throughput screening, hit-to-lead optimization, structure-based design, molecular modelling, synthesis, and biological evaluation. In the cellular target engagement assays, 8h enhances isoproterenol-mediated cyclic adenosine 3',5'-monophosphate (cAMP) production in HEK293 cells overexpressing GRK2. Compound 8h was further evaluated in a human stem cell-derived cardiomyocyte (HSC-CM) contractility assay and potentiated isoproterenol-induced beating rate in HSC-CMs.
Organizational Affiliation:
Discovery Sciences, Janssen Research & Development, L.L.C., Welsh & McKean Roads, Spring House, PA 19477, United States. Electronic address: gxu4@its.jnj.com.