Disulfide-mediated oligomerization of mutant Cu/Zn-superoxide dismutase associated with canine degenerative myelopathy.
Shino, Y., Muraki, N., Kobatake, Y., Kamishina, H., Kato, R., Furukawa, Y.(2024) Protein Sci 33: e5210-e5210
- PubMed: 39548731
- DOI: https://doi.org/10.1002/pro.5210
- Primary Citation of Related Structures:
8ZD5, 8ZD6 - PubMed Abstract:
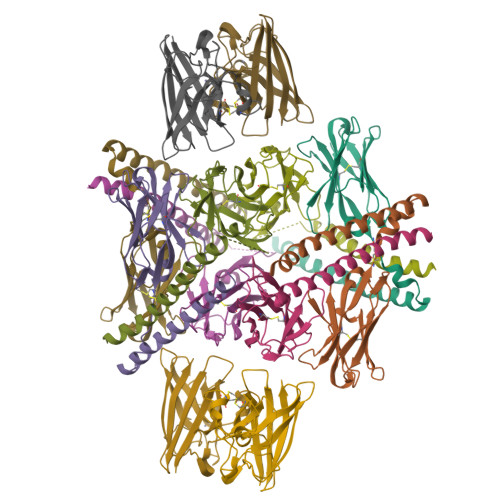
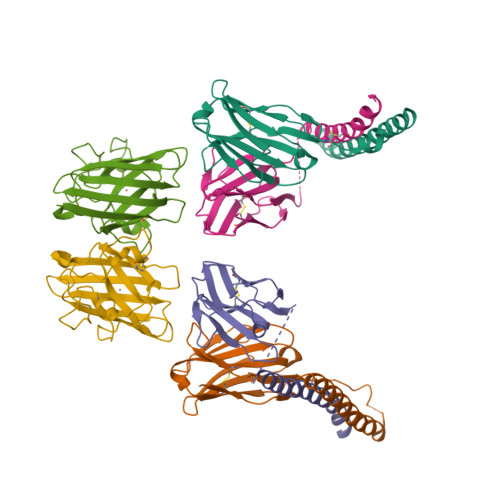
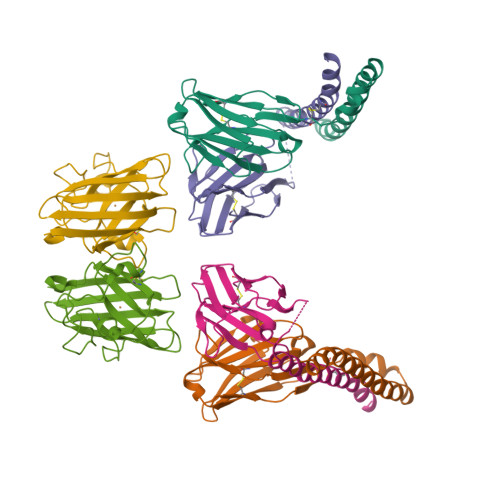
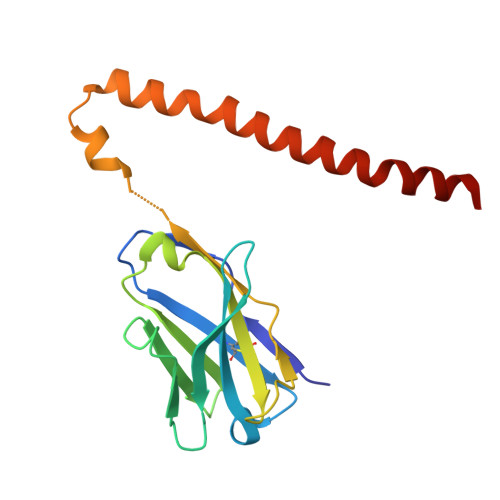
A homozygous E40K mutation in the gene coding canine Cu/Zn-superoxide dismutase (cSOD1) causes degenerative myelopathy (DM) in dogs. A pathological hallmark of DM with the cSOD1 mutation is the aggregation of mutant cSOD1 proteins in neurons. The amino acid substitution E40K disrupts a salt bridge between Glu40 and Lys91 and is considered to destabilize the native state of cSOD1; however, the mechanism by which mutant cSOD1 aggregates remains unclear. Here, we show that mutant cSOD1 losing a copper and zinc ion forms oligomers crosslinked via disulfide bonds. The E40K substitution was found to result in the increased solvent exposure of the Cys7 side chain, which then attacked the disulfide bond (Cys57-Cys146) in cSOD1 to form disulfide-linked oligomers. We also successfully prevented the Cys7 exposure and thus the oligomerization of mutant cSOD1 by a fragment antibody that specifically recognizes the region around the mutation site. The fragment antibody covered the β-plug region, reinforcing the interactions compromised by the E40K substitution and thus contributing to the maintenance of the structural integrity of the β-barrel core of cSOD1. Taken together, we propose that the Cys7 exposure in cSOD1 upon the salt bridge disruption plays a central role in the aggregation mechanism of DM-associated mutant cSOD1.
Organizational Affiliation:
Department of Chemistry, Keio University, Yokohama, Japan.