New Features
Explore Multiple Sequence Alignments in 1D and 3D
12/04
The new 1D-3D Group Alignment Viewer supports exploration of multiple sequence alignments (MSA) at sequence and structure levels for PDB experimental structures and Computed Structure Models (CSMs). Select proteins and/or residue regions from the MSA to view their 3D structures aligned in Mol*. Options to display (or hide) other polymeric chains and ligands are available.
RCSB.org clusters protein entities (PDB experimental structures and CSMs) by sequence identity threshold and UniProt accession. For each cluster, the MSA is calculated using Clustal Omega and displayed in the 1D-3D Group Alignment Viewer using specific color schemes. PDB protein sequence positions are represented in blue if residue was experimentally determined, and in gray if not. CSMs are colored according to their local pLDDT scores.
To access this feature, select the 1D-3D Alignments option from a UniProt Group Summary or Sequence page (for an example, see the Group Summary for O95786). UniProt Group pages are available from Structure Summary pages (in the Entity Groups table) and from Search Results (more). Documentation is available.
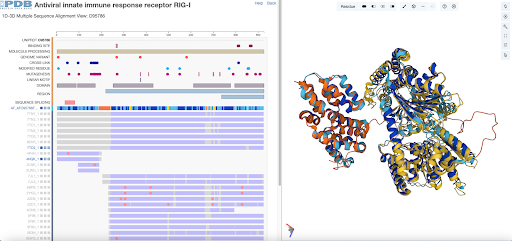 1D-3D Group Alignment View for O95786.
1D-3D Group Alignment View for O95786.













