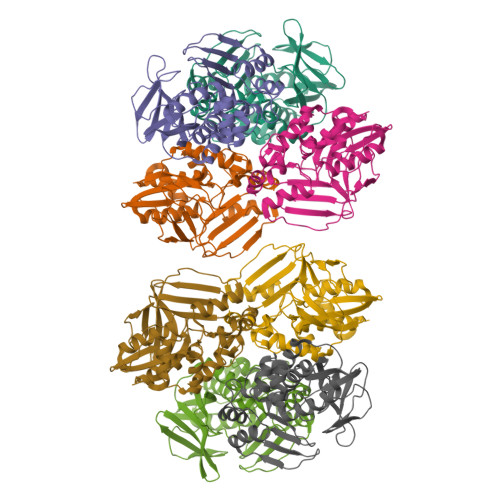
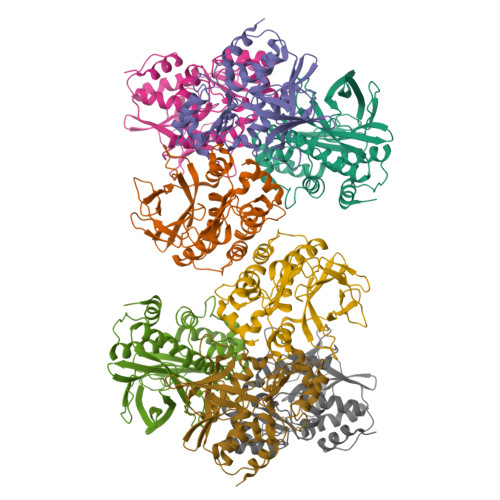
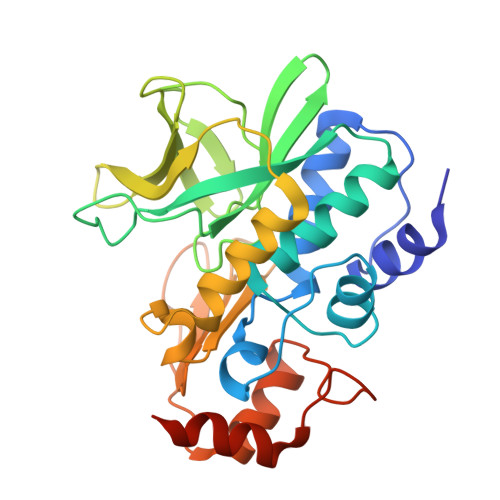
Structure of Arylamine N-Acetyltransferase Reveals a Catalytic Triad
Sinclair, J.C., Sandy, J., Delgoda, R., Sim, E., Noble, M.E.M.(2000) Nat Struct Biol 7: 560
- PubMed: 10876241
- DOI: https://doi.org/10.1038/76783
- Primary Citation of Related Structures:
1E2T - PubMed Abstract:
Enzymes of the arylamine N-acetyltransferase (NAT) family are found in species ranging from Escherichia coli to humans. In humans they are known to be responsible for the acetylation of a number of arylamine and hydrazine drugs, and they are strongly linked to the carcinogenic potentiation of certain foreign substances. In prokaryotes their substrate specificities may vary and members of the gene family have been linked to pathways including amide synthesis during rifamycin production. Here we report the crystal structure at 2.8 A resolution of a representative member of this family from Salmonella typhimurium in the presence and absence of a covalently bound product analog. The structure reveals surprising mechanistic information including the presence of a Cys-His-Asp catalytic triad. The fold can be described in terms of three domains of roughly equal length with the second and third domains linked by an interdomain helix. The first two domains, a helical bundle and a beta-barrel, make up the catalytic triad using a structural motif identical to that of the cysteine protease superfamily.
Organizational Affiliation:
Laboratory of Molecular Biophysics and Oxford Centre for Molecular Sciences, Rex Richards Building, Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, UK.