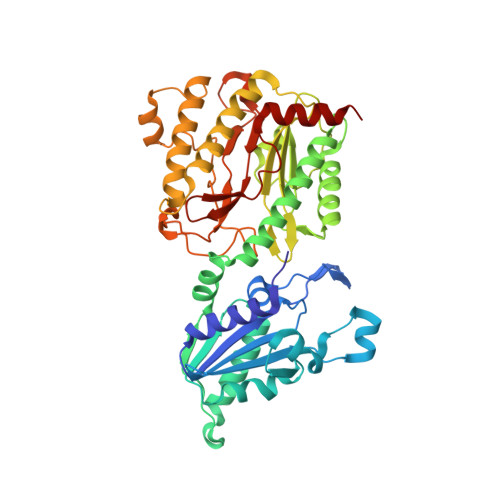
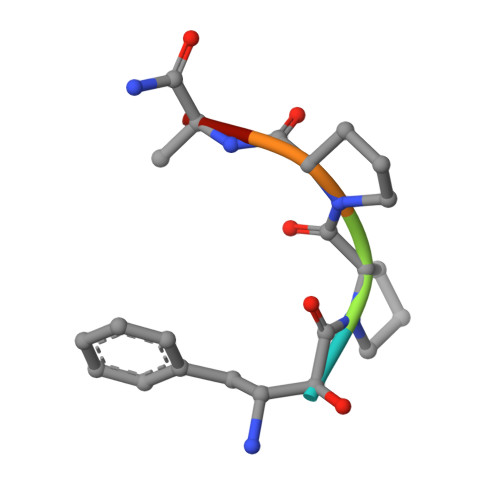
Structure of Escherichia coli aminopeptidase P in complex with the inhibitor apstatin.
Graham, S.C., Maher, M.J., Simmons, W.H., Freeman, H.C., Guss, J.M.(2004) Acta Crystallogr D Biol Crystallogr 60: 1770-1779
- PubMed: 15388923
- DOI: https://doi.org/10.1107/S0907444904018724
- Primary Citation of Related Structures:
1N51 - PubMed Abstract:
Aminopeptidase P (APPro) is a metalloprotease whose active site includes a dinuclear manganese(II) cluster. The enzyme cleaves the N-terminal residue from a polypeptide when the second residue is proline. A complex of Escherichia coli APPro (EcAPPro) with an inhibitor, apstatin [N-(2S,3R)-3-amino-2-hydroxy-4-phenyl-butanoyl-L-prolyl-L-prolyl-L-alaninamide], has been crystallized. Apstatin binds to the active site of EcAPPro with its N-terminal amino group coordinated to one of the two Mn(II) atoms at the metal centre. The apstatin hydroxyl group replaces a hydroxide ion which bridges the two metal atoms in the native enzyme. The first proline residue of apstatin lies in a small hydrophobic cleft. The structure of the apstatin-EcAPPro complex has been refined at 2.3 A resolution with residuals R = 0.179 and R(free) = 0.204. The structure of the complex illustrates how apstatin inhibits APPro and suggests how substrates may bind to the enzyme, but the basis of the proline-specificity remains elusive.
Organizational Affiliation:
School of Molecular and Microbial Biosciences, University of Sydney, NSW 2006, Australia.