The structure at 2.4 A resolution of the protein from gene locus At3g21360, a putative Fe(II)/2-oxoglutarate-dependent enzyme from Arabidopsis thaliana.
Bitto, E., Bingman, C.A., Allard, S.T., Wesenberg, G.E., Aceti, D.J., Wrobel, R.L., Frederick, R.O., Sreenath, H., Vojtik, F.C., Jeon, W.B., Newman, C.S., Primm, J., Sussman, M.R., Fox, B.G., Markley, J.L., Phillips, G.N.(2005) Acta Crystallogr Sect F Struct Biol Cryst Commun 61: 469-472
- PubMed: 16511070
- DOI: https://doi.org/10.1107/S1744309105011565
- Primary Citation of Related Structures:
1Y0Z - PubMed Abstract:
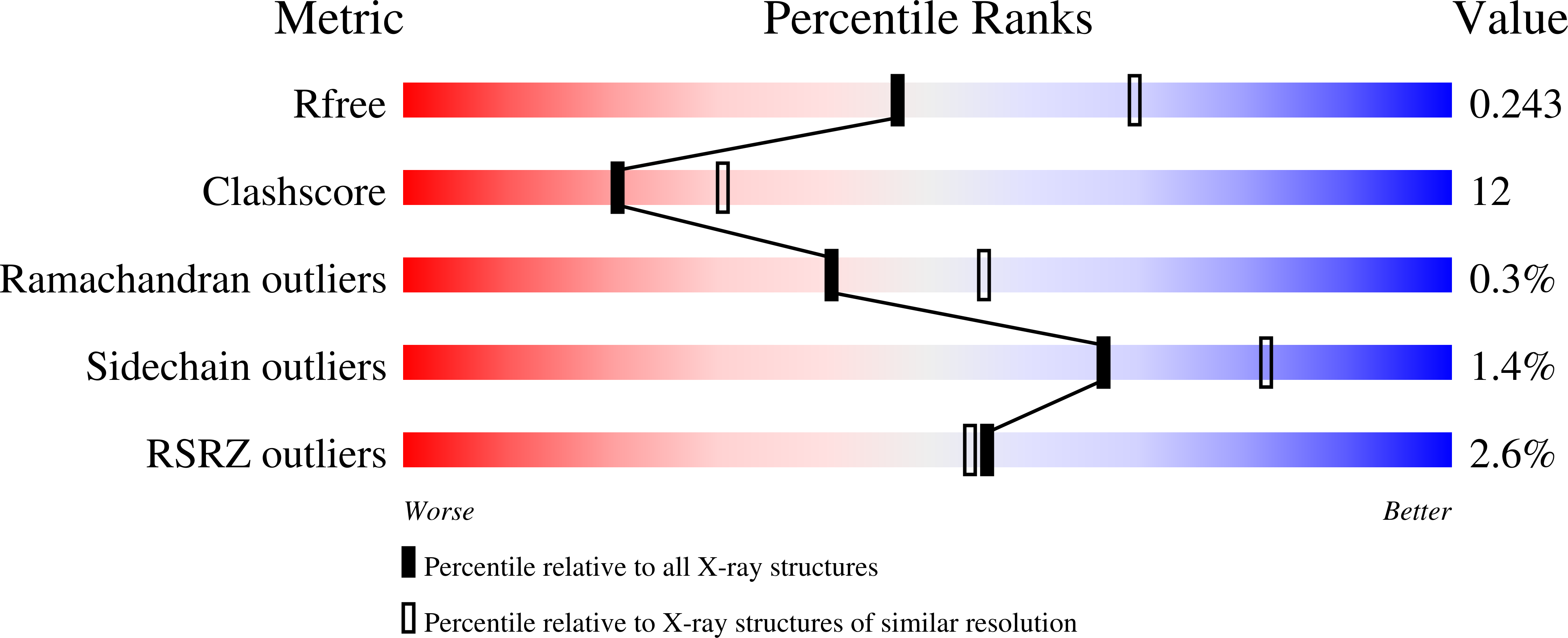
The crystal structure of the gene product of At3g21360 from Arabidopsis thaliana was determined by the single-wavelength anomalous dispersion method and refined to an R factor of 19.3% (Rfree = 24.1%) at 2.4 A resolution. The crystal structure includes two monomers in the asymmetric unit that differ in the conformation of a flexible domain that spans residues 178-230. The crystal structure confirmed that At3g21360 encodes a protein belonging to the clavaminate synthase-like superfamily of iron(II) and 2-oxoglutarate-dependent enzymes. The metal-binding site was defined and is similar to the iron(II) binding sites found in other members of the superfamily.
Organizational Affiliation:
Center for Eukaryotic Structural Genomics, Department of Biochemistry, University of Wisconsin-Madison, USA.