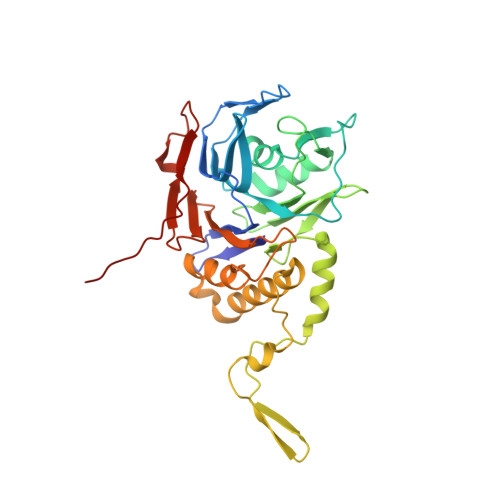
Conjugated Bile Acid Hydrolase is a Tetrameric N-Terminal Thiol Hydrolase with Specific Recognition of its Cholyl But not of its Tauryl Product
Rossocha, M., Schultz-Heienbrok, R., Von Moeller, H., Coleman, J.P., Saenger, W.(2005) Biochemistry 44: 5739
- PubMed: 15823032
- DOI: https://doi.org/10.1021/bi0473206
- Primary Citation of Related Structures:
2BJF, 2BJG - PubMed Abstract:
Bacterial bile salt hydrolases catalyze the degradation of conjugated bile acids in the mammalian gut. The crystal structures of conjugated bile acid hydrolase (CBAH) from Clostridium perfringens as apoenzyme and in complex with taurodeoxycholate that was hydrolyzed to the reaction products taurine and deoxycholate are described here at 2.1 and 1.7 A resolution, respectively. The crystal structures reveal close relationship between CBAH and penicillin V acylase from Bacillus sphaericus. This similarity together with the N-terminal cysteine classifies CBAH as a member of the N-terminal nucleophile (Ntn) hydrolase superfamily. Both crystal structures show an identical homotetrameric organization with dihedral (D(2) or 222) point group symmetry. The structure analysis of C. perfringens CBAH identifies critical residues in catalysis, substrate recognition, and tetramer formation which may serve in further biochemical characterization of bile acid hydrolases.
Organizational Affiliation:
Freie Universität Berlin, Institut für Kristallographie, Takustrasse 6, 14195 Berlin, Germany.