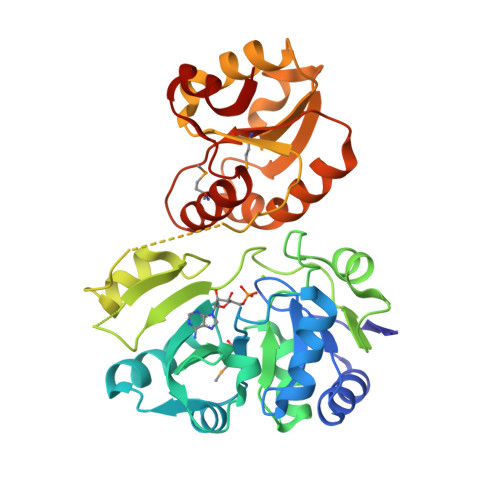
X-ray crystal structure of a hypothetical Sua5 protein from Sulfolobus tokodaii strain 7
Agari, Y., Sato, S., Wakamatsu, T., Bessho, Y., Ebihara, A., Yokoyama, S., Kuramitsu, S., Shinkai, A.(2008) Proteins 70: 1108-1111
- PubMed: 18004774
- DOI: https://doi.org/10.1002/prot.21794
- Primary Citation of Related Structures:
2EQA
Organizational Affiliation:
RIKEN SPring-8 Center, Harima Institute, 1-1-1 Kouto, Sayo, Hyogo 679-5148, Japan.