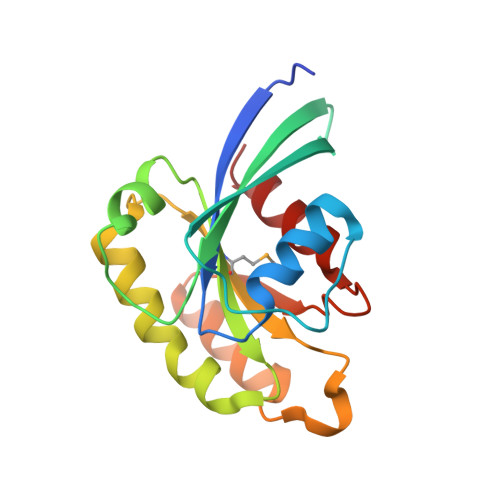
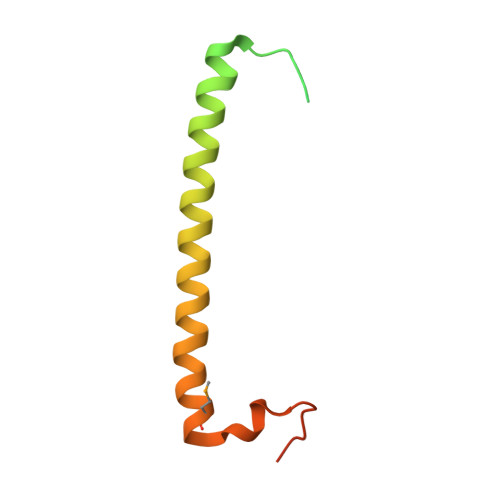
Crystal structure of rab11 in complex with rab11 family interacting protein 2.
Jagoe, W.N., Lindsay, A.J., Read, R.J., McCoy, A.J., McCaffrey, M.W., Khan, A.R.(2006) Structure 14: 1273-1283
- PubMed: 16905101
- DOI: https://doi.org/10.1016/j.str.2006.06.010
- Primary Citation of Related Structures:
2GZD, 2GZH - PubMed Abstract:
The small GTPase Rab11 regulates the recycling of endosomes to the plasma membrane via interactions with the Rab11 family of interacting proteins (FIPs). FIPs contain a highly conserved Rab binding domain (RBD) at their C termini whose structure is unknown. Here, we have determined the crystal structure of the RBD of FIP2 in complex with Rab11(GTP) by single wavelength anomalous diffraction methods. The overall structure is a heterotetramer with dyad symmetry, arranged as a Rab11-(FIP2)2-Rab11 complex. FIP2 forms a central alpha-helical coiled coil, with both helices contributing to the Rab11 binding patch on equivalent and opposite sides of the homodimer. Switch 1 of Rab11 is embedded between the two helices, while switch 2 remains flexible and is peripherally associated with the effector. The complex reveals the structural basis for Rab11 recognition by FIPs and suggests the molecular mechanisms underlying endocytic recycling pathways.
Organizational Affiliation:
School of Biochemistry and Immunology, Trinity College, Dublin 2, Ireland.