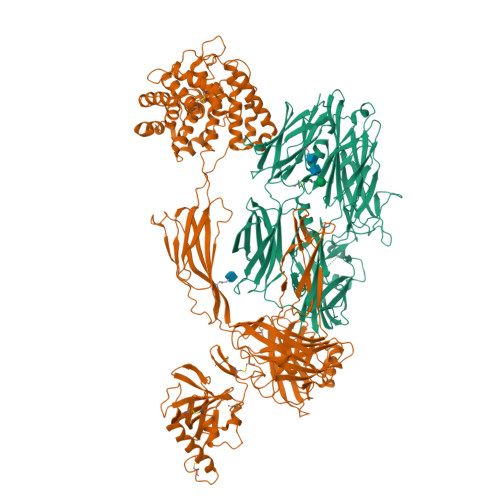
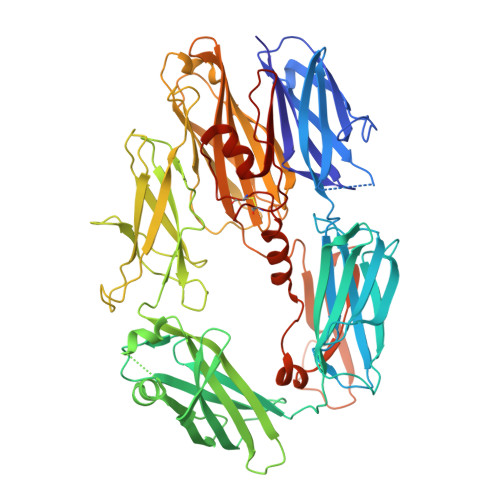
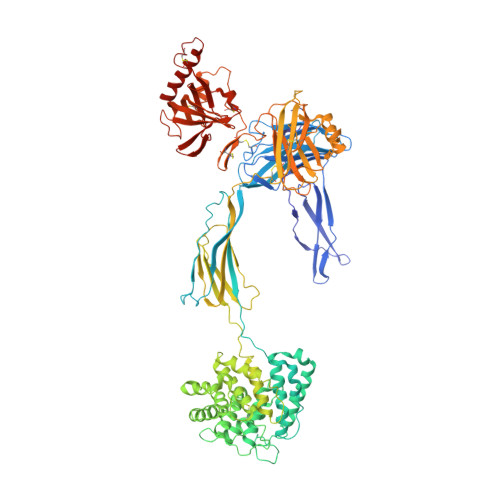
Structure of C3b reveals conformational changes that underlie complement activity.
Janssen, B.J., Christodoulidou, A., McCarthy, A., Lambris, J.D., Gros, P.(2006) Nature 444: 213-216
- PubMed: 17051160
- DOI: https://doi.org/10.1038/nature05172
- Primary Citation of Related Structures:
2I07 - PubMed Abstract:
Resistance to infection and clearance of cell debris in mammals depend on the activation of the complement system, which is an important component of innate and adaptive immunity. Central to the complement system is the activated form of C3, called C3b, which attaches covalently to target surfaces to amplify complement response, label cells for phagocytosis and stimulate the adaptive immune response. C3b consists of 1,560 amino-acid residues and has 12 domains. It binds various proteins and receptors to effect its functions. However, it is not known how C3 changes its conformation into C3b and thereby exposes its many binding sites. Here we present the crystal structure at 4-A resolution of the activated complement protein C3b and describe the conformational rearrangements of the 12 domains that take place upon proteolytic activation. In the activated form the thioester is fully exposed for covalent attachment to target surfaces and is more than 85 A away from the buried site in native C3 (ref. 5). Marked domain rearrangements in the alpha-chain present an altered molecular surface, exposing hidden and cryptic sites that are consistent with known putative binding sites of factor B and several complement regulators. The structural data indicate that the large conformational changes in the proteolytic activation and regulation of C3 take place mainly in the first conversion step, from C3 to C3b. These insights are important for the development of strategies to treat immune disorders that involve complement-mediated inflammation.
Organizational Affiliation:
Crystal and Structural Chemistry, Bijvoet Center for Biomolecular Research, Faculty of Sciences, Utrecht University, Padualaan 8, 3584 CH Utrecht, The Netherlands.