The Structure of an Rnai Polymerase Links RNA Silencing and Transcription.
Salgado, P.S., Koivunen, M.R.L., Makeyev, E.V., Bamford, D.H., Stuart, D.I., Grimes, J.M.(2006) PLoS Biol 4: E434
- PubMed: 17147473
- DOI: https://doi.org/10.1371/journal.pbio.0040434
- Primary Citation of Related Structures:
2J7N, 2J7O - PubMed Abstract:
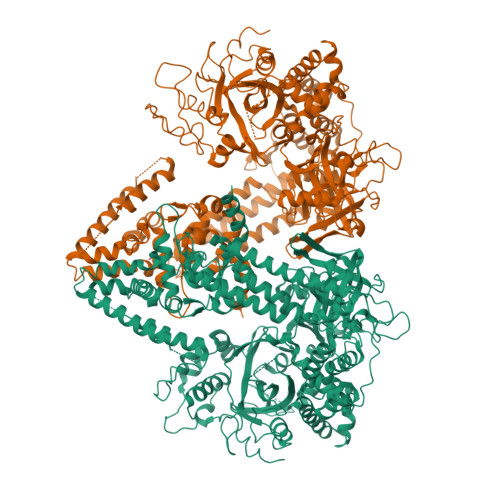
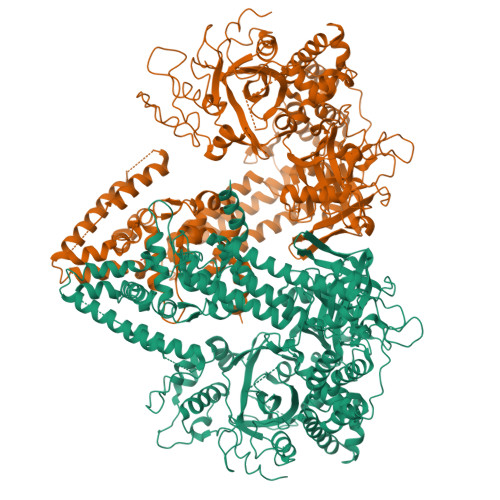
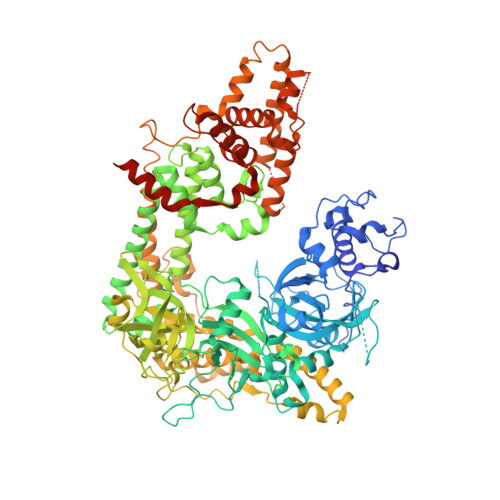
RNA silencing refers to a group of RNA-induced gene-silencing mechanisms that developed early in the eukaryotic lineage, probably for defence against pathogens and regulation of gene expression. In plants, protozoa, fungi, and nematodes, but apparently not insects and vertebrates, it involves a cell-encoded RNA-dependent RNA polymerase (cRdRP) that produces double-stranded RNA triggers from aberrant single-stranded RNA. We report the 2.3-A resolution crystal structure of QDE-1, a cRdRP from Neurospora crassa, and find that it forms a relatively compact dimeric molecule, each subunit of which comprises several domains with, at its core, a catalytic apparatus and protein fold strikingly similar to the catalytic core of the DNA-dependent RNA polymerases responsible for transcription. This evolutionary link between the two enzyme types suggests that aspects of RNA silencing in some organisms may recapitulate transcription/replication pathways functioning in the ancient RNA-based world.
Organizational Affiliation:
Division of Structural Biology, The Henry Wellcome Building for Genomic Medicine, Oxford University, Oxford, United Kingdom.