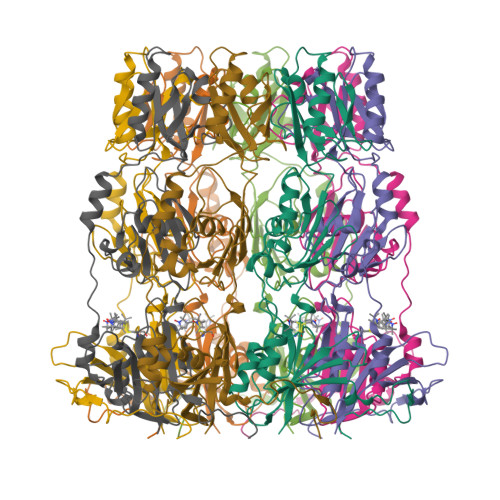
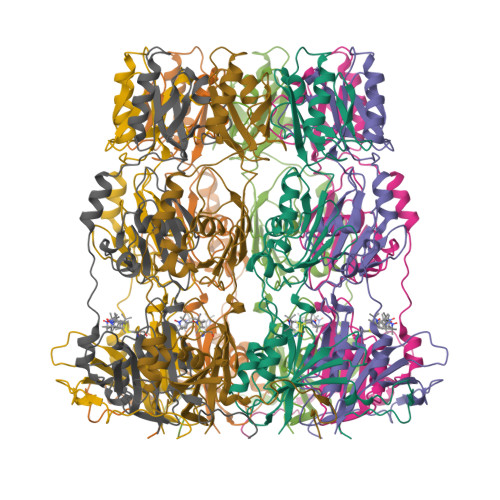
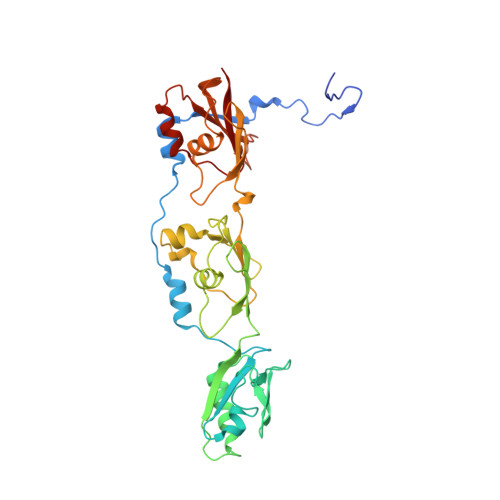
Peldor Distance Fingerprinting of the Octameric Outer-Membrane Protein Wza from Escherichia Coli.
Hagelueken, G., Ingledew, W.J., Huang, H., Petrovic-Stojanovska, B., Whitfield, C., Elmkami, H., Schiemann, O., Naismith, J.H.(2009) Angew Chem Int Ed Engl 48: 2904
- PubMed: 19294709
- DOI: https://doi.org/10.1002/anie.200805758
- Primary Citation of Related Structures:
2W8H, 2W8I - PubMed Abstract:
Distance fingerprinting: Pulsed electron-electron double resonance spectroscopy (PELDOR) is applied to the octameric membrane protein complex Wza of E. coli. The data yielded a detailed distance fingerprint of its periplasmic region that compares favorably to the crystal structure. These results provide the foundation to study conformation changes from interaction with partner proteins.
Organizational Affiliation:
Centre for Biomolecular Sciences, The University of St. Andrews, Fife KY16 9RH, UK.