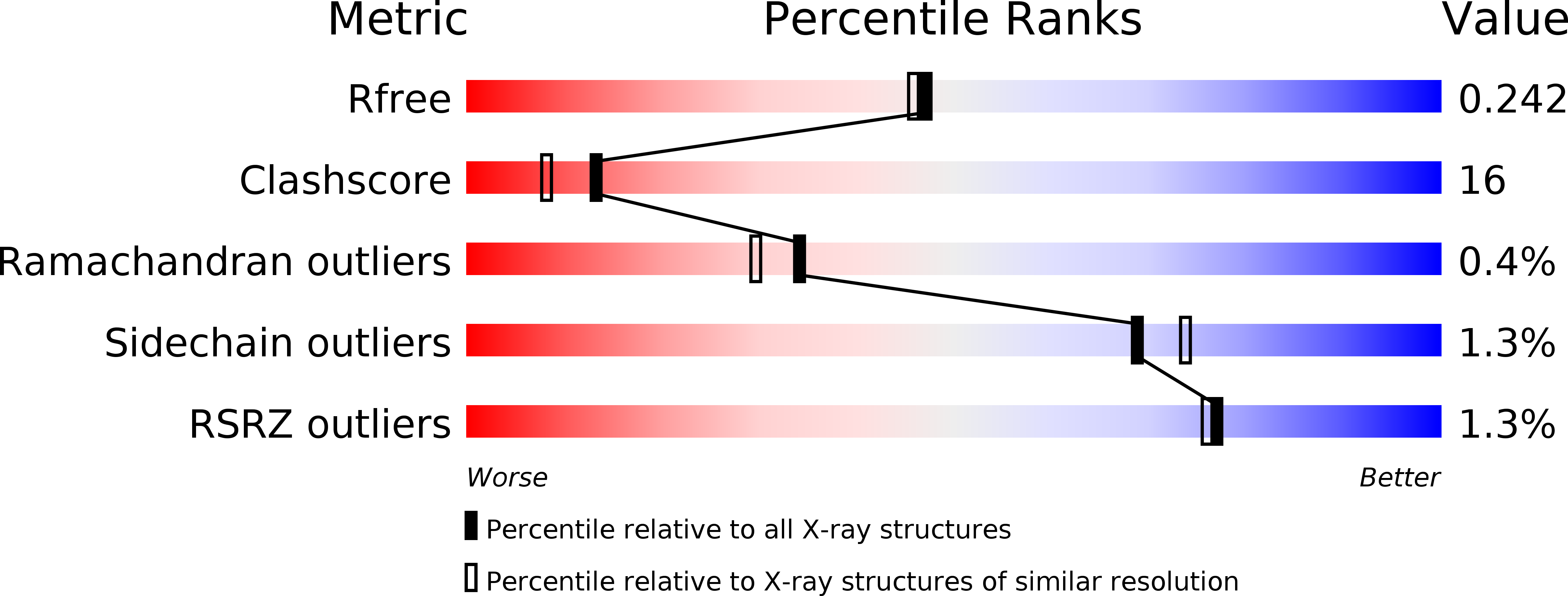
Structures of OppA and PstS from Yersinia pestis indicate variability of interactions with transmembrane domains.
Tanabe, M., Mirza, O., Bertrand, T., Atkins, H.S., Titball, R.W., Iwata, S., Brown, K.A., Byrne, B.(2007) Acta Crystallogr D Biol Crystallogr 63: 1185-1193
- PubMed: 18007034
- DOI: https://doi.org/10.1107/S0907444907048299
- Primary Citation of Related Structures:
2Z22, 2Z23 - PubMed Abstract:
Bacterial ATP-binding cassette (ABC) transport systems couple ATP hydrolysis with the uptake and efflux of a wide range of substances across bacterial membranes. These systems are comprised of transmembrane domains, nucleotide binding domains and, in the case of uptake systems, periplasmic binding proteins responsible for binding and presentation of substrate to the transmembrane domains. In pathogenic bacteria, ABC systems are known to play roles in virulence and pathogenicity and the surface localization of some components has made them attractive targets for both vaccine and anti-infective development. Here, the crystallization of five proteins (OppA, PstS, PiuA, YrbD and CysP) from Yersinia pestis, the causative agent of plague, are reported that diffracted to resolution limits ranging from 1.6 to 5 A. The first crystal structures of ABC system components from Y. pestis, OppA and PstS, are also reported here as complexes with their substrates. Comparisons of these two structures with known structures of related proteins suggest that these proteins possess versatility in substrate recognition and variations in protein-protein interactions with their cognate transmembrane domains.
Organizational Affiliation:
Membrane Protein Crystallography, Division of Molecular Biosciences, Imperial College London, London SW7 2AZ, England.