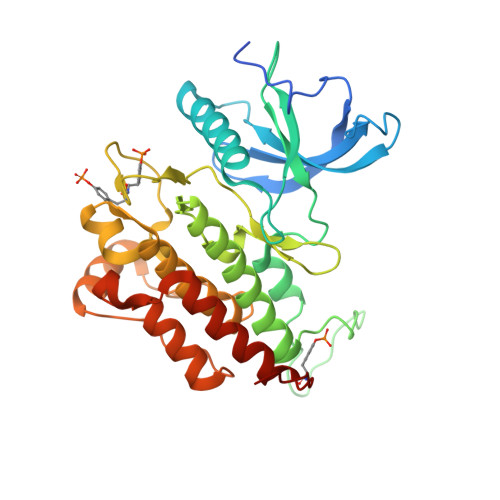
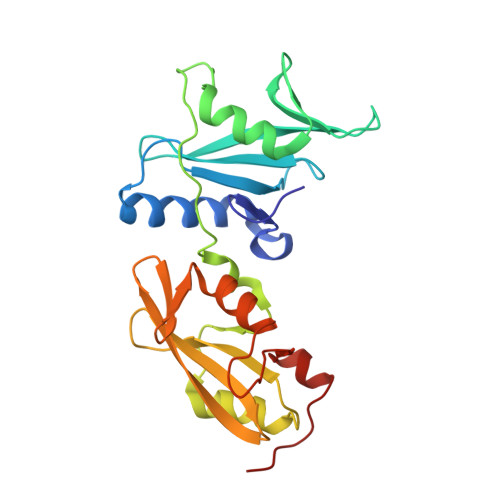
The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site.
Bae, J.H., Lew, E.D., Yuzawa, S., Tome, F., Lax, I., Schlessinger, J.(2009) Cell 138: 514-524
- PubMed: 19665973
- DOI: https://doi.org/10.1016/j.cell.2009.05.028
- Primary Citation of Related Structures:
3GQI, 3GQL - PubMed Abstract:
SH2 domain-mediated interactions represent a crucial step in transmembrane signaling by receptor tyrosine kinases. SH2 domains recognize phosphotyrosine (pY) in the context of particular sequence motifs in receptor phosphorylation sites. However, the modest binding affinity of SH2 domains to pY containing peptides may not account for and likely represents an oversimplified mechanism for regulation of selectivity of signaling pathways in living cells. Here we describe the crystal structure of the activated tyrosine kinase domain of FGFR1 in complex with a phospholipase Cgamma fragment. The structural and biochemical data and experiments with cultured cells show that the selectivity of phospholipase Cgamma binding and signaling via activated FGFR1 are determined by interactions between a secondary binding site on an SH2 domain and a region in FGFR1 kinase domain in a phosphorylation independent manner. These experiments reveal a mechanism for how SH2 domain selectivity is regulated in vivo to mediate a specific cellular process.
Organizational Affiliation:
Department of Pharmacology, Yale University School of Medicine, 333 Cedar Street, New Haven, CT 06520, USA.